Structural Variation and Immune Recognition of the P1.2 Subtype Meningococcal Antigen.
Tzitzilonis, C., Prince, S.M., Collins, R.F., Achtman, M., Feavers, I.M., Maiden, M.C.J., Derrick, J.P.(2005) Proteins 62: 947
- PubMed: 16470851
- DOI: https://doi.org/10.1002/prot.20800
- Primary Citation of Related Structures:
1UWX - PubMed Abstract:
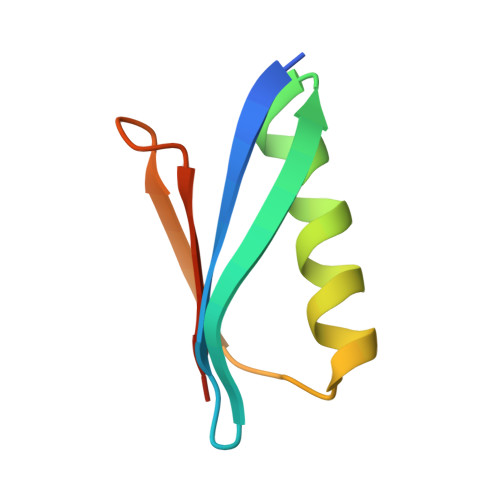
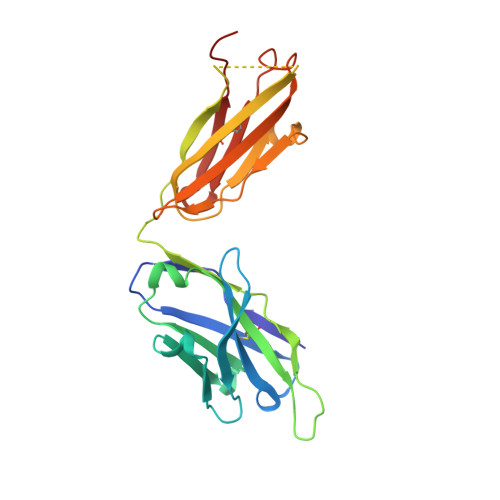
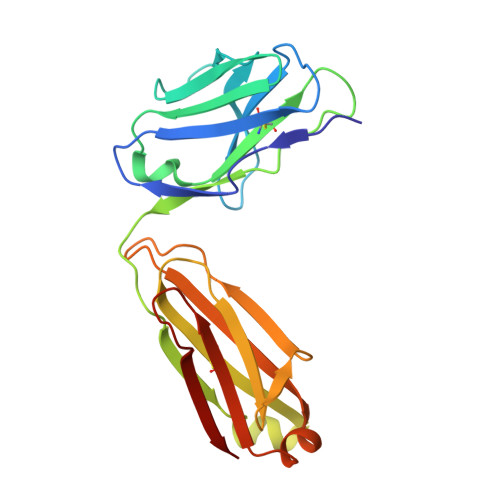
Neisseria meningitidis is a globally important cause of bacterial meningitis and septicemia. No comprehensive antimeningococcal vaccine is available, largely as a consequence of the high sequence diversity of those surface proteins that could function as components of a vaccine. One such component is the protein PorA, a major surface porin of this Gram-negative organism that has been used in a number of experimental and licensed vaccines. Here we describe a series of experiments designed to investigate the consequences for antibody recognition of sequence diversity within a PorA antigen. The binding of a 14-residue peptide, corresponding to the P1.2 subtype antigen, to the MN16C13F4 monoclonal antibody was sensitive to mutation of five out of the six residues within the epitope sequence. The crystal structure of the antibody Fab fragment, determined in complex with the peptide antigen, shows a remarkably hydrophobic binding site and interactions between the antigen and antibody are dominated by apolar residues. Nine intrachain hydrogen bonds are formed within the antigen which maintain the beta-hairpin conformation of the peptide. These hydrogen bonds involve residues that are highly conserved amongst different P1.2 sequence variants, suggesting that some positions may be conserved for structural reasons in these highly polymorphic regions. The sensitivity of antibody recognition of the antigen towards mutation provides a structural explanation for the widespread sequence variation seen in different PorA sequences in this region. Single point mutations are sufficient to remove binding capability, providing a rationale for the manner in which different meningococcal PorA escape variants arise.
- Faculty of Life Sciences, The University of Manchester, Manchester, United Kingdom.
Organizational Affiliation: