Refinement of the C222(1) crystal form of oxidized uteroglobin at 1.34 A resolution.
Morize, I., Surcouf, E., Vaney, M.C., Epelboin, Y., Buehner, M., Fridlansky, F., Milgrom, E., Mornon, J.P.(1987) J Mol Biology 194: 725-739
- PubMed: 3656405
- DOI: https://doi.org/10.1016/0022-2836(87)90250-6
- Primary Citation of Related Structures:
1UTG - PubMed Abstract:
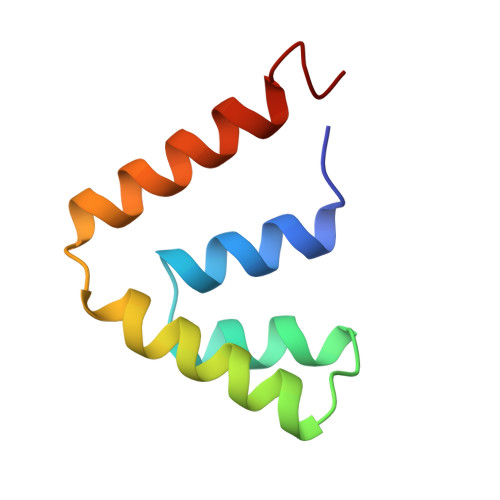
The structure of uteroglobin, a progesterone binding protein from rabbit uterine fluid, was determined and refined at 1.34 A resolution to a conventional R-factor of 0.229. The accuracy of the co-ordinates is estimated to be 0.15 A. The isotropic temperature factor of individual atoms was refined and its average value is 11.9 A2 for the 548 non-hydrogen atoms of the protein monomer. A total of 83 water molecules was located in difference electron density maps and refined, first using a constant occupancy factor of 1 and then variable occupancy, the final (Q) being 0.63. The mean temperature factor of the water oxygen atoms is 26.4 A2. Uteroglobin is a dimer and its secondary structure consists of four alpha-helices per monomer that align in an anti-parallel fashion. There is one beta-turn between helix 2 and helix 3 (Lys26 to Glu29); 76% of the residues are part of the alpha-helices. In the core of the dimeric protein molecule, between the two monomers that are held together by two disulfide bridges, we have observed a closed cavity. Its length is 15.6 A and its width is 9 A; 14 water molecules could be positioned inside. In the "bottom" part of the protein, near the C terminus, we have observed a smaller cavity, occupied by two water molecules. The calculation of the molecular surface revealed four surface pockets whose possible functional implications are discussed below.
- Laboratoire de Minéralogie-Cristallographie associé au CNRS, Universités Paris, France.
Organizational Affiliation: