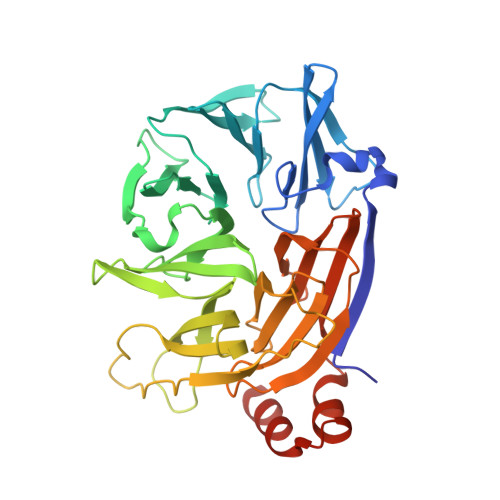
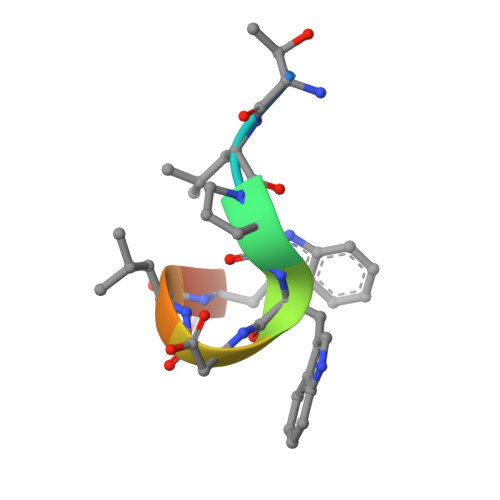
Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller.
Miele, A.E., Watson, P.J., Evans, P.R., Traub, L.M., Owen, D.J.(2004) Nat Struct Mol Biol 11: 242-248
- PubMed: 14981508
- DOI: https://doi.org/10.1038/nsmb736
- Primary Citation of Related Structures:
1UTC - PubMed Abstract:
During the assembly of clathrin-coated vesicles, many peripheral membrane proteins, including the amphiphysins, use LLDLD-type clathrin-box motifs to interact with the N-terminal beta-propeller domain (TD) of clathrin. The 2.3 A-resolution structure of the clathrin TD in complex with a TLPWDLWTT peptide from amphiphysin 1 delineates a second clathrin-binding motif, PWXXW (the W box), that binds at a site on the TD remote from the clathrin box-binding site. The presence of both sequence motifs within the unstructured region of the amphiphysins allows them to bind more tightly to free TDs than do other endocytic proteins that contain only clathrin-box motifs. This property, along with the propensity of the N-terminal BAR domain to bind curved membranes, will preferentially localize amphiphysin and its partner, dynamin, to the periphery of invaginated clathrin lattices.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.
Organizational Affiliation: