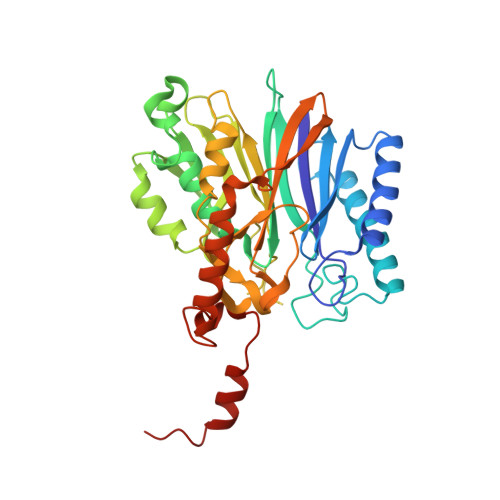
The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Nishimasu, H., Fushinobu, S., Shoun, H., Wakagi, T.(2004) Structure 12: 949-959
- PubMed: 15274916
- DOI: https://doi.org/10.1016/j.str.2004.03.026
- Primary Citation of Related Structures:
1UMG - PubMed Abstract:
As the first structure of the novel class of fructose-1,6-bisphosphatase (FBPase) present in thermophilic archaea, we solved the crystal structure of the ST0318 gene product (St-Fbp) of Sulfolobus tokodaii strain 7. The St-Fbp structure comprises a homooctamer of the 422 point-group. The protein folds as a four-layer alpha-beta-beta-alpha sandwich with a novel topology, which is completely different from the sugar phosphatase fold. The structure contains an unhydrolyzed FBP molecule in the open-keto form, as well as four hexacoordinated magnesium ions around the 1-phosphoryl group of FBP. The arrangement of the catalytic side chains and metal ligands is consistent with the three-metal ion assisted catalysis proposed for conventional FBPases. The structure provides an insight into the structural basis of the strict substrate specificity of St-Fbp.
- Department of Biotechnology, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.
Organizational Affiliation: