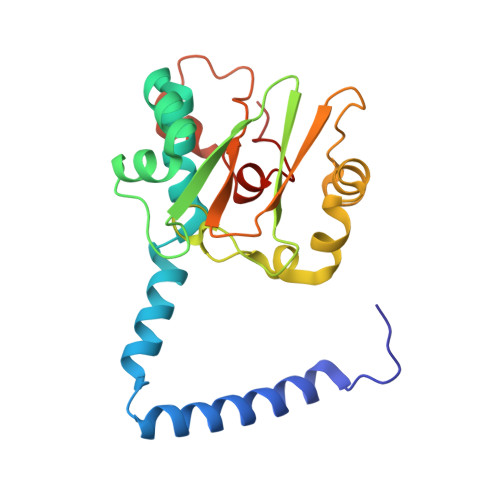
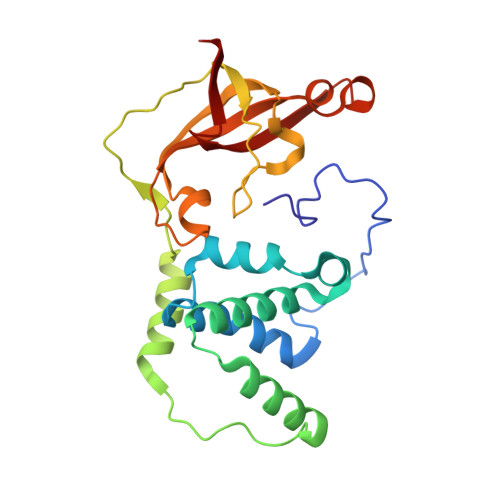
Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Miyanaga, A., Fushinobu, S., Ito, K., Shoun, H., Wakagi, T.(2004) Eur J Biochem 271: 429-438
- PubMed: 14717710
- DOI: https://doi.org/10.1046/j.1432-1033.2003.03943.x
- Primary Citation of Related Structures:
1UGP, 1UGQ, 1UGR, 1UGS - PubMed Abstract:
Mutants of a cobalt-containing nitrile hydratase (NHase, EC 4.2.1.84) from Pseudonocardia thermophila JCM 3095 involved in substrate binding, catalysis and formation of the active center were constructed, and their characteristics and crystal structures were investigated. As expected from the structure of the substrate binding pocket, the wild-type enzyme showed significantly lower K(m) and K(i) values for aromatic substrates and inhibitors, respectively, than aliphatic ones. In the crystal structure of a complex with an inhibitor (n-butyric acid) the hydroxyl group of betaTyr68 formed hydrogen bonds with both n-butyric acid and alphaSer112, which is located in the active center. The betaY68F mutant showed an elevated K(m) value and a significantly decreased k(cat) value. The apoenzyme, which contains no detectable cobalt atom, was prepared from Escherichia coli cells grown in medium without cobalt ions. It showed no detectable activity. A disulfide bond between alphaCys108 and alphaCys113 was formed in the apoenzyme structure. In the highly conserved sequence motif in the cysteine cluster region, two positions are exclusively conserved in cobalt-containing or iron-containing nitrile hydratases. Two mutants (alphaT109S and alphaY114T) were constructed, each residue being replaced with an iron-containing one. The alphaT109S mutant showed similar characteristics to the wild-type enzyme. However, the alphaY114T mutant showed a very low cobalt content and catalytic activity compared with the wild-type enzyme, and oxidative modifications of alphaCys111 and alphaCys113 residues were not observed. The alphaTyr114 residue may be involved in the interaction with the nitrile hydratase activator protein of P. thermophila.
- Department of Biotechnology, The University of Tokyo, Japan.
Organizational Affiliation: