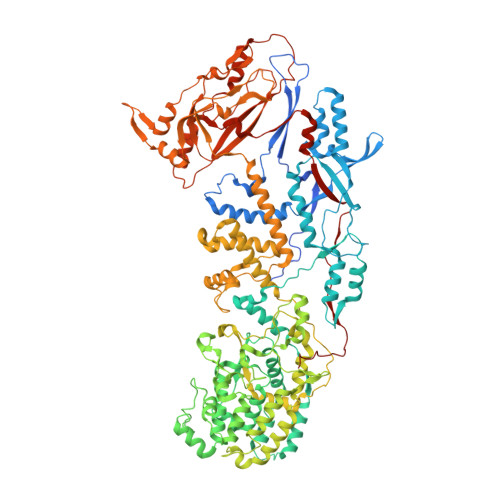
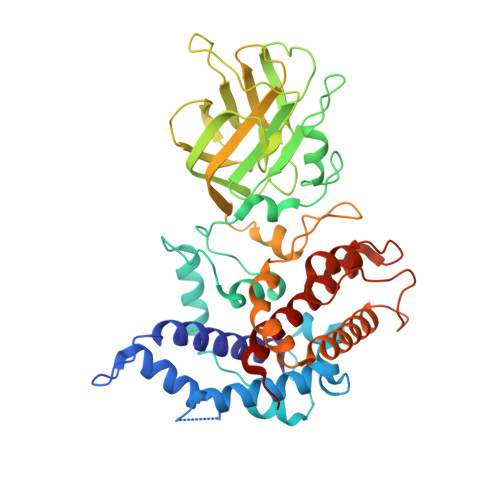
The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Nakagawa, A., Miyazaki, N., Taka, J., Naitow, H., Ogawa, A., Fujimoto, Z., Mizuno, H., Higashi, T., Watanabe, Y., Omura, T., Cheng, R.H., Tsukihara, T.(2003) Structure 11: 1227-1238
- PubMed: 14527391
- DOI: https://doi.org/10.1016/j.str.2003.08.012
- Primary Citation of Related Structures:
1UF2 - PubMed Abstract:
Rice dwarf virus (RDV), the causal agent of rice dwarf disease, is a member of the genus Phytoreovirus in the family Reoviridae. RDV is a double-shelled virus with a molecular mass of approximately 70 million Dalton. This virus is widely prevalent and is one of the viruses that cause the most economic damage in many Asian countries. The atomic structure of RDV was determined at 3.5 A resolution by X-ray crystallography. The double-shelled structure consists of two different proteins, the core protein P3 and the outer shell protein P8. The atomic structure shows structural and electrostatic complementarities between both homologous (P3-P3 and P8-P8) and heterologous (P3-P8) interactions, as well as overall conformational changes found in P3-P3 dimer caused by the insertion of amino-terminal loop regions of one of the P3 protein into the other. These interactions suggest how the 900 protein components are built into a higher-ordered virus core structure.
- Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita, Osaka 565-0871, Japan. atsushi@protein.osaka-u.ac.jp
Organizational Affiliation: