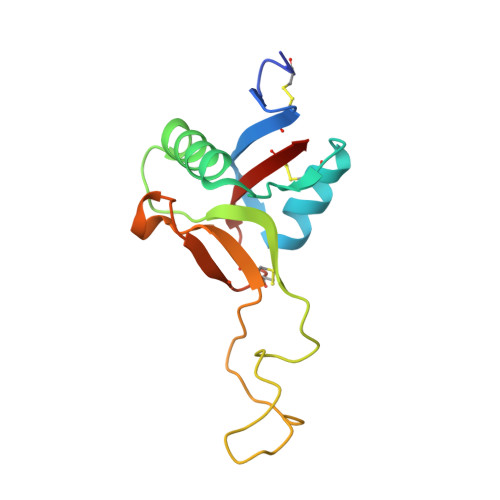
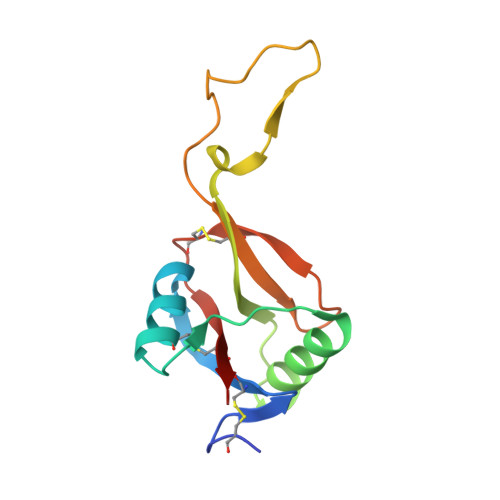
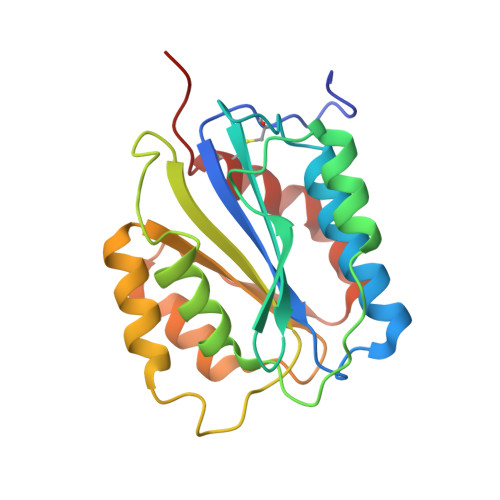
Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin. Insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins.
Maita, N., Nishio, K., Nishimoto, E., Matsui, T., Shikamoto, Y., Morita, T., Sadler, J.E., Mizuno, H.(2003) J Biological Chem 278: 37777-37781
- PubMed: 12851390
- DOI: https://doi.org/10.1074/jbc.M305566200
- Primary Citation of Related Structures:
1UEX - PubMed Abstract:
Bitiscetin, a platelet adhesion inducer isolated from venom of the snake Bitis arietans, activates the binding of the von Willebrand factor (VWF) A1 domain to glycoprotein Ib (GPIb) in vitro. This activation requires the formation of a bitiscetin-VWF A1 complex, suggesting an allosteric mechanism of action. Here, we report the crystal structure of bitiscetin-VWF A1 domain complex solved at 2.85 A. In the complex structure, helix alpha5 of VWF A1 domain lies on a concave depression on bitiscetin, and binding sites are located at both ends of the depression. The binding sites correspond well with those proposed previously based on alanine-scanning mutagenesis (Matsui, T., Hamako, J., Matsushita, T., Nakayama, T., Fujimura, Y., and Titani, K. (2002) Biochemistry 41, 7939-7946). Against our expectations, the structure of the VWF A1 domain bound to bitiscetin does not differ significantly from the structure of the free A1 domain. These results are similar to the case of botrocetin, another snake-derived inducer of platelet aggregation, although the binding modes of botrocetin and bitiscetin are different. The modeled structure of the ternary bitiscetin-VWF A1-GPIb complex suggests that an electropositive surface of bitiscetin may interact with a favorably positioned anionic region of GPIb. These results suggest that snake venom proteins induce VWF A1-GPIbalpha binding by interacting with both proteins, and not by causing conformational changes in VWF A1.
- Department of Biochemistry, National Institute of Agrobiological Sciences, Tsukuba, Ibaraki 305-8602, Japan.
Organizational Affiliation: