Identification of a Novel Family of Proteins in Snake Venoms: Purification and Structural Characterization of Nawaprin from Naja nigricollis Snake Venom
Torres, A.M., Wong, H.Y., Desai, M., Moochhala, S., Kuchel, P.W., Kini, R.M.(2003) J Biological Chem 278: 40097-40104
- PubMed: 12878611
- DOI: https://doi.org/10.1074/jbc.M305322200
- Primary Citation of Related Structures:
1UDK - PubMed Abstract:
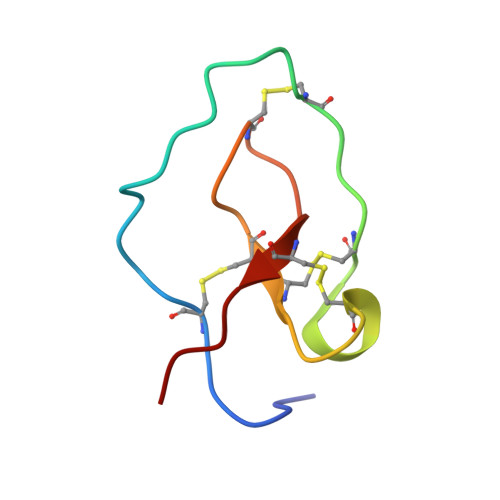
The three-dimensional structure of nawaprin has been determined by nuclear magnetic resonance spectroscopy. This 51-amino acid residue peptide was isolated from the venom of the spitting cobra, Naja nigricollis, and is the first member of a new family of snake venom proteins referred to as waprins. Nawaprin is relatively flat and disc-like in shape, characterized by a spiral backbone configuration that forms outer and inner circular segments. The two circular segments are held together by four disulfide bonds, three of which are clustered at the base of the molecule. The inner segment contains a short antiparallel beta-sheet, whereas the outer segment is devoid of secondary structures except for a small turn or 310 helix. The structure of nawaprin is very similar to elafin, a human leukocyte elastase-specific inhibitor. Although substantial parts of the nawaprin molecule are well defined, the tips of the outer and inner circular segments, which are hypothesized to be critical for binding interactions, are apparently disordered, similar to that found in elafin. The amino acid residues in these important regions in nawaprin are different from those in elafin, suggesting that nawaprin is not an elastase-specific inhibitor and therefore has a different function in the snake venom.
- School of Molecular and Microbial Biosciences, University of Sydney, Sydney, New South Wales 2006, Australia.
Organizational Affiliation: