Bovine thrombin complexed with an uncleavable analog of residues 7-19 of fibrinogen A alpha: geometry of the catalytic triad and interactions of the P1', P2', and P3' substrate residues.
Martin, P.D., Malkowski, M.G., DiMaio, J., Konishi, Y., Ni, F., Edwards, B.F.(1996) Biochemistry 35: 13030-13039
- PubMed: 8855938
- DOI: https://doi.org/10.1021/bi960656y
- Primary Citation of Related Structures:
1UCY - PubMed Abstract:
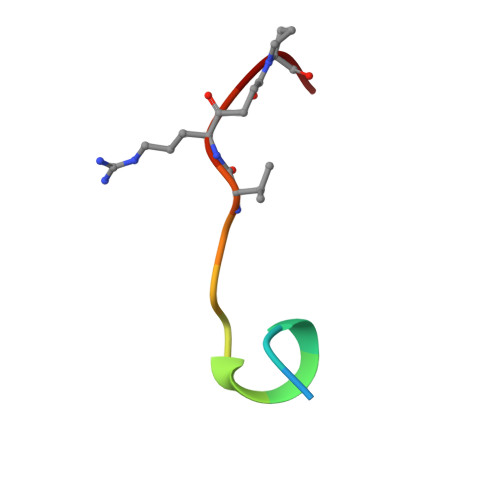
The crystal structure of the noncovalent complex of bovine thrombin and a fibrinogen-A alpha tridecapeptide substrate analog, G17 psi, in which the scissile bond amide nitrogen of Gly-17f has been replaced by a methylene carbon, has been determined at 2.3 A resolution with an R factor of 17.1%. The geometry of the active site indicates that the crystal structure is a close model of the true Michaelis complex. The three independently determined thrombin/G17 psi complexes in the crystal asymmetric unit reveal novel interactions for the P2' and P3' residues-Pro-18f and Arg-19f, respectively-on the carboxyl-terminal side of the scissile bond and confirm previously observed interactions of the P1 (Arg-16f) through P10 (Asp-7f) positions on the amino-terminal side. The thrombin S2' binding site for Pro-18f, as observed in all three complexes, differs from that predicted by modeling studies and is notable for including two carbonyl oxygens of the thrombin main chain. Arg-19f occupies two binding sites on thrombin, S3'A and S3'B, which have dramatically different placements for the arginyl side chain and carboxyl terminus.
- Department of Biochemistry, Wayne State University, Detroit, Michigan 48201, USA.
Organizational Affiliation: