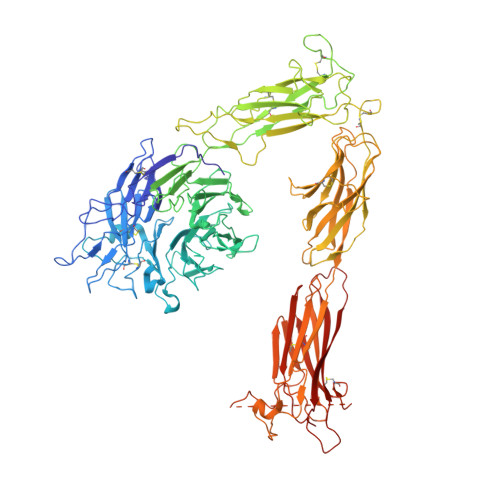
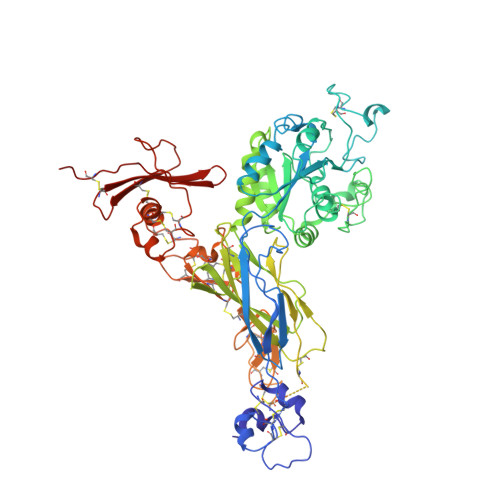
A novel adaptation of the integrin PSI domain revealed from its crystal structure.
Xiong, J.P., Stehle, T., Goodman, S.L., Arnaout, M.A.(2004) J Biological Chem 279: 40252-40254
- PubMed: 15299032
- DOI: https://doi.org/10.1074/jbc.C400362200
- Primary Citation of Related Structures:
1U8C - PubMed Abstract:
Integrin beta-subunits contain an N-terminal PSI (for plexin-semaphorin-integrin) domain that contributes to integrin activation and harbors the PI(A) alloantigen associated with immune thrombocytopenias and susceptibility to sudden cardiac death. Here we report the crystal structure of PSI in the context of the crystallized alphaVbeta3 ectodomain. The integrin PSI forms a two-stranded antiparallel beta-sheet flanked by two short helices; its long interstrand loop houses Pl(A) and may face the EGF2 domain. The integrin PSI contains four cysteine pairs connected in a 1-4, 2-8, 3-6, 5-7 pattern. An unexpected feature of the structure is that the final, eighth cysteine is located C-terminal to the Ig-like hybrid domain and is thus separated by the hybrid domain from the other seven cysteines of PSI. This architecture may be relevant to the evolution of integrins and should help refine the current models of integrin activation.
- Structural Biology Program, Leukocyte Biology and Inflammation Program, Renal Unit.
Organizational Affiliation: