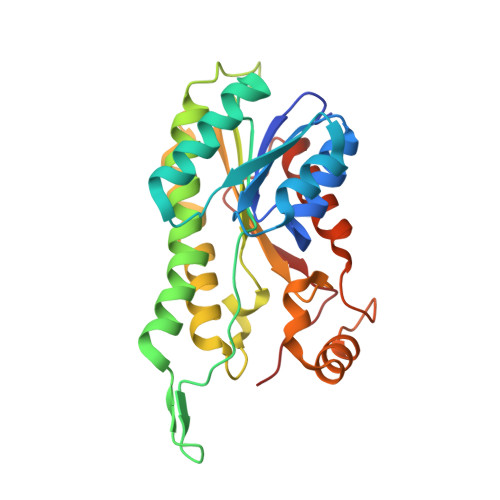
Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics
Kissinger, C.R., Rejto, P.A., Pelletier, L.A., Thomson, J.A., Showalter, R.E., Abreo, M.A., Agree, C.S., Margosiak, S., Meng, J.J., Aust, R.M., Vanderpool, D., Li, B., Tempczyk-Russell, A., Villafranca, J.E.(2004) J Mol Biology 342: 943-952
- PubMed: 15342248
- DOI: https://doi.org/10.1016/j.jmb.2004.07.071
- Primary Citation of Related Structures:
1U7T - PubMed Abstract:
The enzyme 17beta-hydroxysteroid dehydrogenase type 10 (HSD10), also known as amyloid beta-peptide-binding alcohol dehydrogenase (ABAD), has been implicated in the development of Alzheimer's disease. This protein, a member of the short-chain dehydrogenase/reductase family of enzymes, has been shown to bind beta-amyloid and to participate in beta-amyloid neurotoxicity. We have determined the crystal structure of human ABAD/HSD10 complexed with NAD(+) and an inhibitory small molecule. The inhibitor occupies the substrate-binding site and forms a covalent adduct with the NAD(+) cofactor. The crystal structure provides a basis for the design of potent, highly specific ABAD/HSD10 inhibitors with potential application in the treatment of Alzheimer's disease.
- Pfizer-La Jolla, 10777 Science Center Dr., San Diego, CA 92121, USA.
Organizational Affiliation: