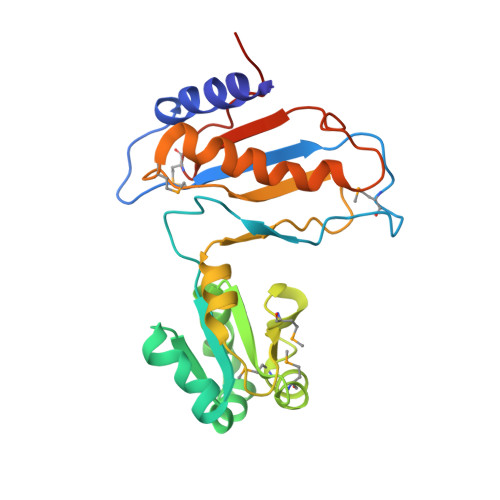
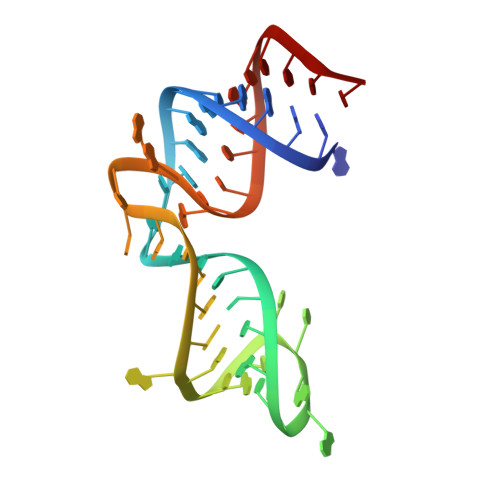
Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA.
Nevskaya, N., Tishchenko, S., Gabdoulkhakov, A., Nikonova, E., Nikonov, O., Nikulin, A., Platonova, O., Garber, M., Nikonov, S., Piendl, W.(2005) Nucleic Acids Res 33: 478-485
- PubMed: 15659579
- DOI: https://doi.org/10.1093/nar/gki194
- Primary Citation of Related Structures:
1U63 - PubMed Abstract:
The RNA-binding ability of ribosomal protein L1 is of profound interest since the protein has a dual function as a ribosomal protein binding rRNA and as a translational repressor binding its mRNA. Here, we report the crystal structure of ribosomal protein L1 in complex with a specific fragment of its mRNA and compare it with the structure of L1 in complex with a specific fragment of 23S rRNA determined earlier. In both complexes, a strongly conserved RNA structural motif is involved in L1 binding through a conserved network of RNA-protein H-bonds inaccessible to the solvent. These interactions should be responsible for specific recognition between the protein and RNA. A large number of additional non-conserved RNA-protein H-bonds stabilizes both complexes. The added contribution of these non-conserved H-bonds makes the ribosomal complex much more stable than the regulatory one.
- Institute of Protein Research, Russian Academy of Sciences 142290 Pushchino, Moscow region, Russia.
Organizational Affiliation: