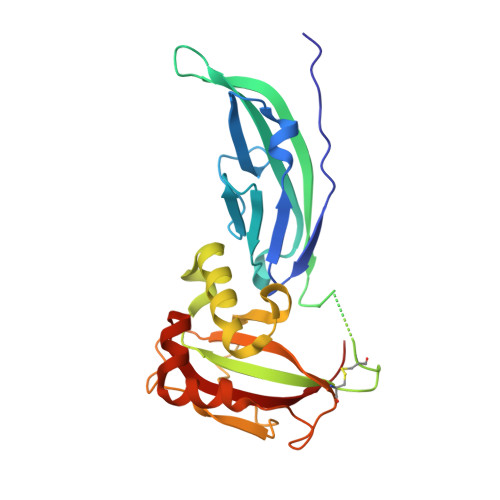
The Structure of the N-terminal Region of Murine Skeletal Muscle {alpha}-Dystroglycan Discloses a Modular Architecture
Bozic, D., Sciandra, F., Lamba, D., Brancaccio, A.(2004) J Biological Chem 279: 44812-44816
- PubMed: 15326183
- DOI: https://doi.org/10.1074/jbc.C400353200
- Primary Citation of Related Structures:
1U2C - PubMed Abstract:
Dystroglycan (DG) is a cell surface receptor consisting of two subunits: alpha-dystroglycan, extracellular and highly glycosylated, and beta-dystroglycan, spanning the cell membrane. It is a pivotal member of the dystrophin-glycoprotein complex and is involved in a wide variety of important cellular processes such as the stabilization of the muscle fiber sarcolemma or the clustering of acetylcholine receptors. We report the 2.3-A resolution crystal structure of the murine skeletal muscle N-terminal alpha-DG region, which confirms the presence of two autonomous domains; the first finally identified as an Ig-like and the second resembling ribosomal RNA-binding proteins. Solid-phase laminin binding assays show the occurrence of protein-protein type of interactions involving the Ig-like domain of alpha-DG.
- Biochemisches Institut der Universität Zürich, Zürich 8044, Switzerland. damir.bozic@mat.ethz.ch
Organizational Affiliation: