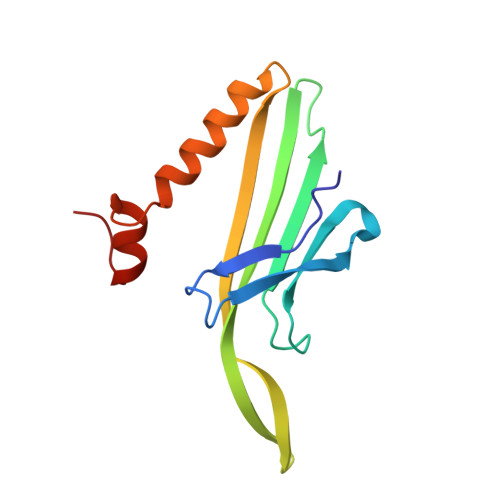
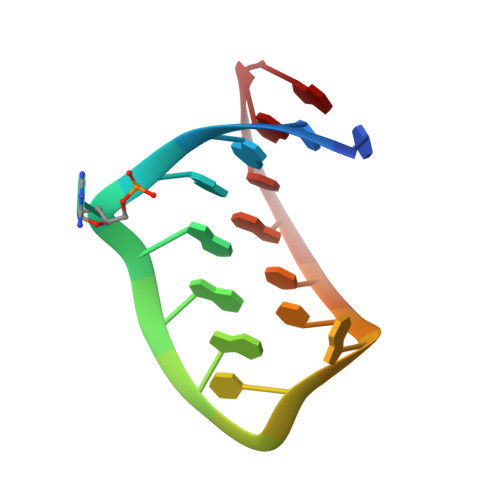
The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions.
Horn, W.T., Convery, M.A., Stonehouse, N.J., Adams, C.J., Liljas, L., Phillips, S.E., Stockley, P.G.(2004) RNA 10: 1776-1782
- PubMed: 15496523
- DOI: https://doi.org/10.1261/rna.7710304
- Primary Citation of Related Structures:
1U1Y - PubMed Abstract:
We have determined the structure to 2.8 A of an RNA aptamer (F5), containing 2'-deoxy-2-aminopurine (2AP) at the -10 position, complexed with MS2 coat protein by soaking the RNA into precrystallised MS2 capsids. The -10 position of the RNA is an important determinant of binding affinity for coat protein. Adenine at this position in other RNA stem-loops makes three hydrogen bonds to protein functional groups. Substituting 2AP for the -10 adenine in the F5 aptamer yields an RNA with the highest yet reported affinity for coat protein. The refined X-ray structure shows that the 2AP base makes an additional hydrogen bond to the protein compared to adenine that is presumably the principal origin of the increased affinity. There are also slight changes in phosphate backbone positions compared to unmodified F5 that probably also contribute to affinity. Such phosphate movements are common in structures of RNAs bound to the MS2 T = 3 protein shell and highlight problems for de novo design of RNA binding ligands.
- Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, United Kingdom.
Organizational Affiliation: