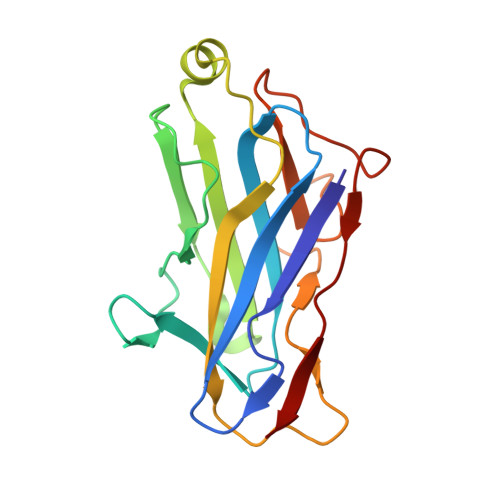
Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements
Noach, I., Frolow, F., Jakoby, H., Rosenheck, S., Shimon, L.J.W., Lamed, R., Bayer, E.A.(2005) J Mol Biology 348: 1-12
- PubMed: 15808849
- DOI: https://doi.org/10.1016/j.jmb.2005.02.024
- Primary Citation of Related Structures:
1QZN, 1TYJ - PubMed Abstract:
The incorporation of enzymes into the multi-enzyme cellulosome complex and its anchoring to the bacterial cell surface are dictated by a set of binding interactions between two complementary protein modules: the cohesin and the dockerin. In this work, the X-ray crystal structure of a type-II cohesin from scaffoldin A of Bacteroides cellulosolvens has been determined to a resolution of 1.6 angstroms using molecular replacement. The type-II B. cellulosolvens cohesin (Bc-cohesin-II) is the first detailed description of a crystal structure for a type-II cohesin, and its features were compared with the known type-I cohesins from Clostridium thermocellum and Clostridium cellulolyticum (Ct-cohesin-I and Cc-cohesin-I, respectively). The overall jelly-roll topology of the type-II Bc-cohesin is very similar to that observed for the type-I cohesins with three additional secondary structures: an alpha-helix and two "beta-flaps" that disrupt the normal course of a beta-strand. In addition, beta-strand 5 is elevated by approximately 4 angstroms on the surface of the molecule, relative to the type-I Ct and Cc-cohesins. Like its type-I analogue, the hydrophobic/aromatic core of Bc-cohesin-II comprises an upper and lower core, but an additional aromatic patch and conserved tryptophan at the crown of the molecule serves to stabilize the alpha-helix of the type-II cohesin. Comparison of Bc-cohesin-II with the known type-I cohesin-dockerin heterodimer suggests that each of the additional secondary structural elements assumes a flanking position relative to the putative dockerin-binding surface. The raised ridge formed by beta-strand 5 confers additional distinctive topographic features to the proposed binding interface that collectively distinguish between the type-II and type-I cohesins.
- Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: