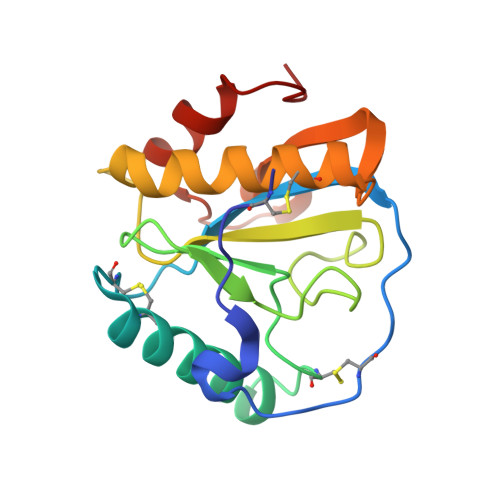
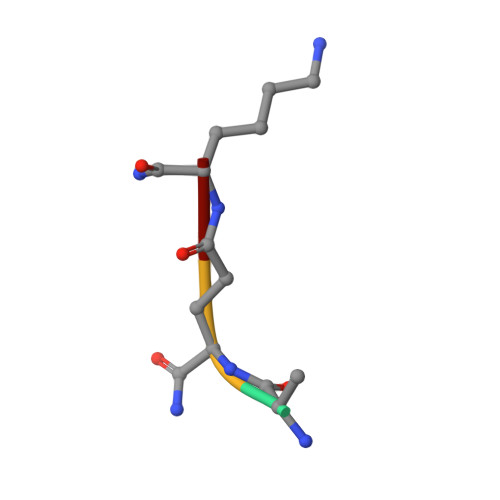
Structural basis for peptidoglycan binding by peptidoglycan recognition proteins
Guan, R., Roychowdhury, A., Ember, B., Kumar, S., Boons, G.-A., Mariuzza, R.A.(2004) Proc Natl Acad Sci U S A 101: 17168-17173
- PubMed: 15572450
- DOI: https://doi.org/10.1073/pnas.0407856101
- Primary Citation of Related Structures:
1TWQ - PubMed Abstract:
Peptidoglycan (PGN) recognition proteins (PGRPs) are pattern-recognition receptors of the innate immune system that bind and, in some cases, hydrolyze bacterial PGNs. We determined the crystal structure, at 2.30-A resolution, of the C-terminal PGN-binding domain of human PGRP-Ialpha in complex with a muramyl tripeptide representing the core of lysine-type PGNs from Gram-positive bacteria. The peptide stem of the ligand is buried at the deep end of a long binding groove, with N-acetylmuramic acid situated in the middle of the groove, whose shallow end can accommodate a linked N-acetylglucosamine. Although most interactions are with the peptide, the glycan moiety also seems to be essential for specific recognition by PGRPs. Conservation of key PGN-contacting residues shows that all PGRPs employ this basic PGN-binding mode. The structure pinpoints variable residues that likely mediate discrimination between lysine- and diaminopimelic acid-type PGNs. We also propose a mechanism for PGN hydrolysis by Zn(2+)-containing PGRPs.
- Center for Advanced Research in Biotechnology, W. M. Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Rockville, MD 20850, USA.
Organizational Affiliation: