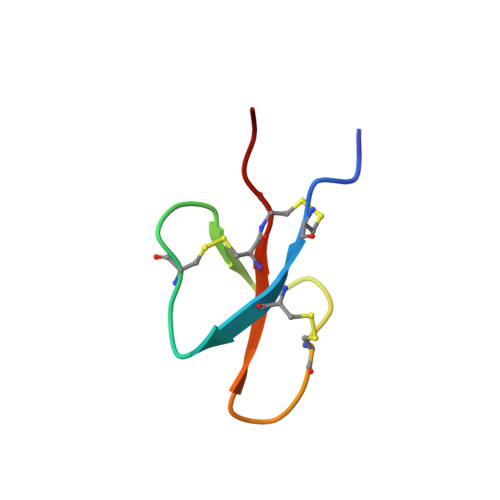
Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Jing, W., Hunter, H.N., Tanabe, H., Ouellette, A.J., Vogel, H.J.(2004) Biochemistry 43: 15759-15766
- PubMed: 15595831
- DOI: https://doi.org/10.1021/bi048645p
- Primary Citation of Related Structures:
1TV0 - PubMed Abstract:
Mammalian defensins are abundant antimicrobial peptides that contribute to host defense. They are characterized by several conserved amino acids, including six invariant cysteine residues which form three intramolecular disulfide bonds and stabilize the tertiary structure. Cryptdin-4 (Crp4), a mouse alpha-defensin with potent in vitro bactericidal activity, has a primary structure distinct from all known alpha-defensins in that its polypeptide backbone uniquely lacks three residues between Cys(IV) and Cys(V). NMR diffusion experiments showed that Crp4 is monomeric in solution, and its three-dimensional solution structure, determined by two-dimensional proton NMR, consists of a triple-stranded antiparallel beta-sheet with the beta-strands joined to each other by a series of tight turns and a beta-hairpin. However, the overall beta-sheet content in Crp4 is lower than that of other alpha-defensin structures, while the shape and orientation of the Crp4 beta-hairpin also differ from those of other alpha-defensin structures. These structural characteristics combined with the high overall cationicity of Crp4 may contribute to its broad bactericidal spectrum and membrane disruptive activity.
- Structural Biological Research Group, Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada.
Organizational Affiliation: