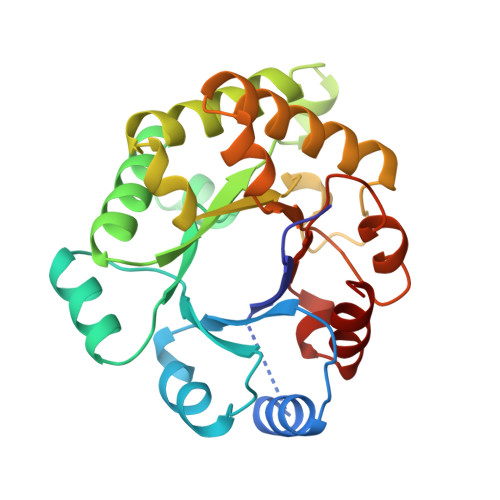
Three new crystal structures of point mutation variants of monoTIM: conformational flexibility of loop-1, loop-4 and loop-8.
Borchert, T.V., Kishan, K.V., Zeelen, J.P., Schliebs, W., Thanki, N., Abagyan, R., Jaenicke, R., Wierenga, R.K.(1995) Structure 3: 669-679
- PubMed: 8591044
- DOI: https://doi.org/10.1016/s0969-2126(01)00202-7
- Primary Citation of Related Structures:
1MSS, 1TTI, 1TTJ - PubMed Abstract:
Wild-type triosephosphate isomerase (TIM) is a very stable dimeric enzyme. This dimer can be converted into a stable monomeric protein (monoTIM) by replacing the 15-residue interface loop (loop-3) by a shorter, 8-residue, loop. The crystal structure of monoTIM shows that two active-site loops (loop-1 and loop-4), which are at the dimer interface in wild-type TIM, have acquired rather different structural properties. Nevertheless, monoTIM has residual catalytic activity. Three new structures of variants of monoTIM are presented, a double-point mutant crystallized in the presence and absence of bound inhibitor, and a single-point mutant in the presence of a different inhibitor. These new structures show large structural variability for the active-site loops, loop-1, loop-4 and loop-8. In the structures with inhibitor bound, the catalytic lysine (Lys13 in loop-1) and the catalytic histidine (His95 in loop-4) adopt conformations similar to those observed in wild-type TIM, but very different from the monoTIM structure. The residual catalytic activity of monoTIM can now be rationalized. In the presence of substrate analogues the active-site loops, loop-1, loop-4 and loop-8, as well as the catalytic residues, adopt conformations similar to those seen in the wild-type protein. These loops lack conformational flexibility in wild-type TIM. The data suggest that the rigidity of these loops in wild-type TIM, resulting from subunit-subunit contacts at the dimer interface, is important for optimal catalysis.
- EMBL, Heidelberg, Germany.
Organizational Affiliation: