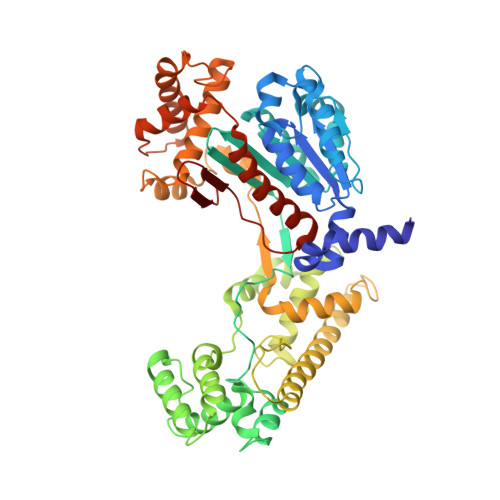
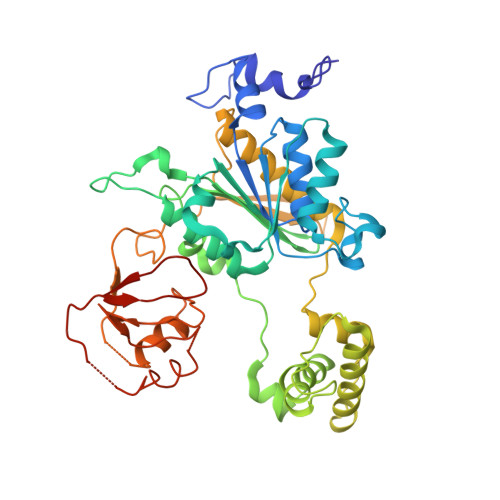
A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Huang, D.T., Miller, D.W., Mathew, R., Cassell, R., Holton, J.M., Roussel, M.F., Schulman, B.A.(2004) Nat Struct Mol Biol 11: 927-935
- PubMed: 15361859
- DOI: https://doi.org/10.1038/nsmb826
- Primary Citation of Related Structures:
1TT5 - PubMed Abstract:
Ubiquitin-like proteins (UBLs) such as NEDD8 are transferred to their targets by distinct, parallel, multienzyme cascades that involve the sequential action of E1, E2 and E3 enzymes. How do enzymes within a particular UBL conjugation cascade interact with each other? We report here that the unique N-terminal sequence of NEDD8's E2, Ubc12, selectively recruits NEDD8's E1 to promote thioester formation between Ubc12 and NEDD8. A peptide corresponding to Ubc12's N terminus (Ubc12N26) specifically binds and inhibits NEDD8's E1, the heterodimeric APPBP1-UBA3 complex. The structure of APPBP1-UBA3- Ubc12N26 reveals conserved Ubc12 residues docking in a groove generated by loops conserved in UBA3s but not other E1s. These data explain why the Ubc12-UBA3 interaction is unique to the NEDD8 pathway. These studies define a novel mechanism for E1-E2 interaction and show how enzymes within a particular UBL conjugation cascade can be tethered together by unique protein-protein interactions emanating from their common structural scaffolds.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, USA.
Organizational Affiliation: