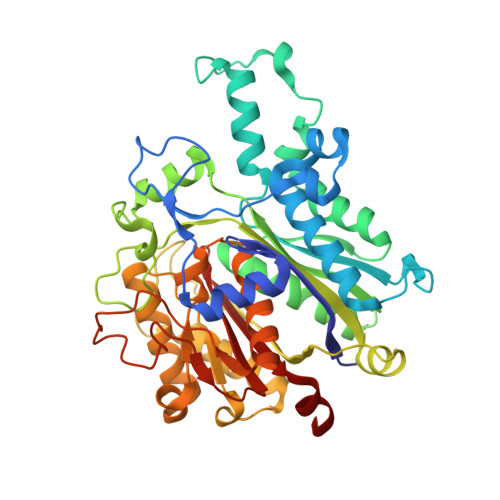
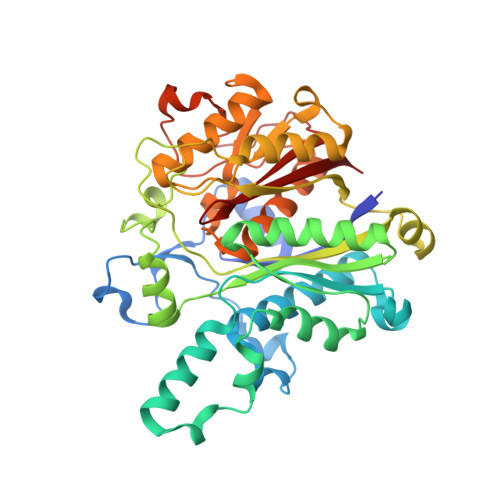
An antibiotic factory caught in action.
Keatinge-Clay, A.T., Maltby, D.A., Medzihradszky, K.F., Khosla, C., Stroud, R.M.(2004) Nat Struct Mol Biol 11: 888-893
- PubMed: 15286722
- DOI: https://doi.org/10.1038/nsmb808
- Primary Citation of Related Structures:
1TQY - PubMed Abstract:
The synthesis of aromatic polyketides, such as actinorhodin, tetracycline and doxorubicin, begins with the formation of a polyketide chain. In type II polyketide synthases (PKSs), chains are polymerized by the heterodimeric ketosynthase-chain length factor (KS-CLF). Here we present the 2.0-A structure of the actinorhodin KS-CLF, which shows polyketides being elongated inside an amphipathic tunnel approximately 17 A in length at the heterodimer interface. The structure resolves many of the questions about the roles of KS and CLF. Although CLF regulates chain length, it does not have an active site; KS must catalyze both chain initiation and elongation. We provide evidence that the first cyclization of the polyketide occurs within the KS-CLF tunnel. The mechanistic details of this central PKS polymerase could guide biosynthetic chemists in designing new pharmaceuticals and polymers.
- Graduate Group in Biophysics, University of California San Francisco, San Francisco, California 94107-2240, USA.
Organizational Affiliation: