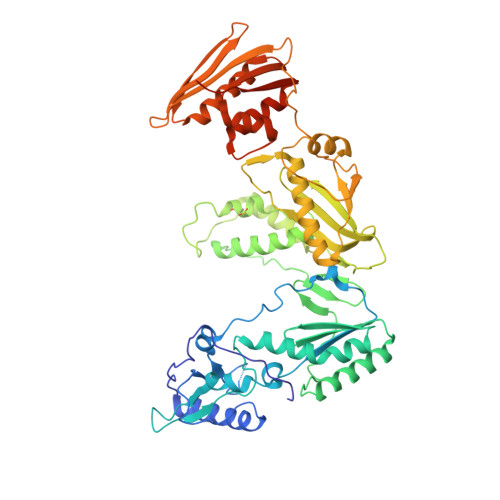
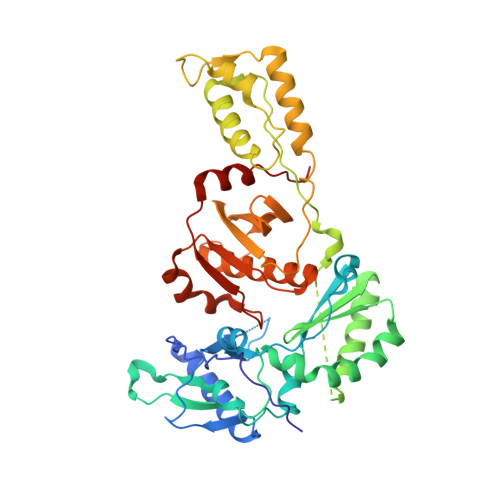
Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
Freeman, G.A., Andrews Iii, C.W., Hopkins, A.L., Lowell, G.S., Schaller, L.T., Cowan, J.R., Gonzales, S.S., Koszalka, G.W., Hazen, R.J., Boone, L.R., Ferris, R.G., Creech, K.L., Roberts, G.B., Short, S.A., Weaver, K., Reynolds, D.J., Milton, J., Ren, J., Stuart, D.I., Stammers, D.K., Chan, J.H.(2004) J Med Chem 47: 5923-5936
- PubMed: 15537347
- DOI: https://doi.org/10.1021/jm040072r
- Primary Citation of Related Structures:
1TKX - PubMed Abstract:
HIV-1 nonnucleoside reverse transcriptase inhibitors (NNRTIs) are part of the combination therapy currently used to treat HIV infection. The features of a new NNRTI drug for HIV treatment must include selective potent activity against both wild-type virus as well as against mutant virus that have been selected by use of current antiretroviral treatment regimens. Based on analogy with known HIV-1 NNRTI inhibitors and modeling studies utilizing the X-ray crystal structure of inhibitors bound in the HIV-1 RT, a series of substituted 2-quinolones was synthesized and evaluated as HIV-1 inhibitors.
- GlaxoSmithKline Research and Development, 5 Moore Drive, Research Triangle Park, North Carolina 27709, USA. Andy.A.Freeman@gsk.com
Organizational Affiliation: