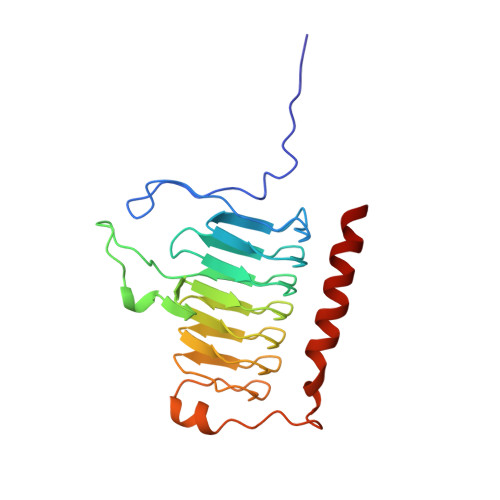
A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.
Kisker, C., Schindelin, H., Alber, B.E., Ferry, J.G., Rees, D.C.(1996) EMBO J 15: 2323-2330
- PubMed: 8665839
- Primary Citation of Related Structures:
1THJ - PubMed Abstract:
A carbonic anhydrase from the thermophilic archaeon Methanosarcina thermophila that exhibits no significant sequence similarity to known carbonic anhydrases has recently been characterized. Here we present the structure of this enzyme, which adopts a left-handed parallel beta-helix fold. This fold is of particular interest since it contains only left-handed crossover connections between the parallel beta-strands, which so far have been observed very infrequently. The active form of the enzyme is a trimer with three zinc-containing active sites, each located at the interface between two monomers. While the arrangement of active site groups differs between this enzyme and the carbonic anhydrases from higher vertebrates, there are structural similarities in the zinc coordination environment, suggestive of convergent evolution dictated by the chemical requirements for catalysis of the same reaction. Based on sequence similarities, the structure of this enzyme is the prototype of a new class of carbonic anhydrases with representatives in all three phylogenetic domains of life.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125, USA. kisker@citray.caltech.edu
Organizational Affiliation: