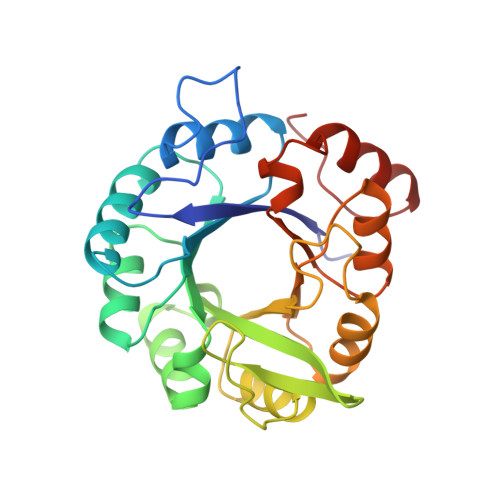
Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion.
Lang, D.A., Obmolova, G., Thoma, R., Kirschner, K., Sterner, R., Wilmanns, M.(2000) Science 289: 1546-1550
- PubMed: 10968789
- DOI: https://doi.org/10.1126/science.289.5484.1546
- Primary Citation of Related Structures:
1QO2, 1THF - PubMed Abstract:
The atomic structures of two proteins in the histidine biosynthesis pathway consist of beta/alpha barrels with a twofold repeat pattern. It is likely that these proteins evolved by twofold gene duplication and gene fusion from a common half-barrel ancestor. These ancestral domains are not visible as independent domains in the extant proteins but can be inferred from a combination of sequence and structural analysis. The detection of subdomain structures may be useful in efforts to search genome sequences for functionally and structurally related proteins.
- European Molecular Biology Laboratory (EMBL) Hamburg Outstation, EMBL c/o Deutsches Elektronen- Synchrotron (DESY), Notkestrasse 85, D-22603 Hamburg, Germany.
Organizational Affiliation: