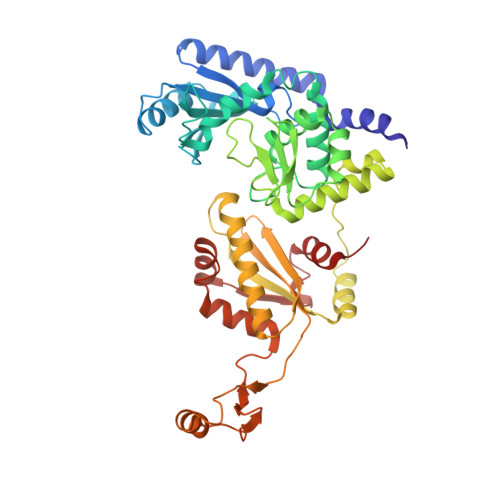
Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template.
Xiong, Y., Steitz, T.A.(2004) Nature 430: 640-645
- PubMed: 15295590
- DOI: https://doi.org/10.1038/nature02711
- Primary Citation of Related Structures:
1SZ1, 1TFW, 1TFY - PubMed Abstract:
Transfer RNA nucleotidyltransferases (CCA-adding enzymes) are responsible for the maturation or repair of the functional 3' end of tRNAs by means of the addition of the essential nucleotides CCA. However, it is unclear how tRNA nucleotidyltransferases polymerize CCA onto the 3' terminus of immature tRNAs without using a nucleic acid template. Here we describe the crystal structure of the Archaeoglobus fulgidus tRNA nucleotidyltransferase in complex with tRNA. We also present ternary complexes of this enzyme with both RNA duplex mimics of the tRNA acceptor stem that terminate with the nucleotides C74 or C75, as well as the appropriate incoming nucleoside 5'-triphosphates. A single nucleotide-binding pocket exists whose specificity for both CTP and ATP is determined by the protein side chain of Arg 224 and backbone phosphates of the tRNA, which are non-complementary to and thus exclude UTP and GTP. Discrimination between CTP or ATP at a given addition step and at termination arises from changes in the size and shape of the nucleotide binding site that is progressively altered by the elongating 3' end of the tRNA.
- Department of Molecular Biophysics and Biochemistry,Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: