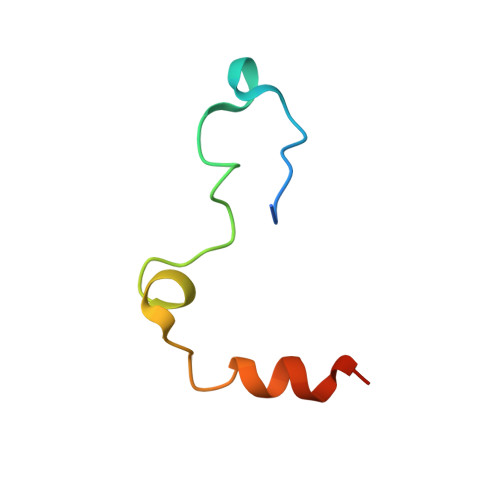
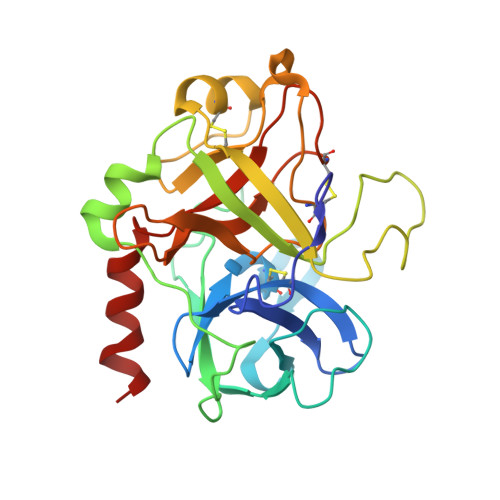
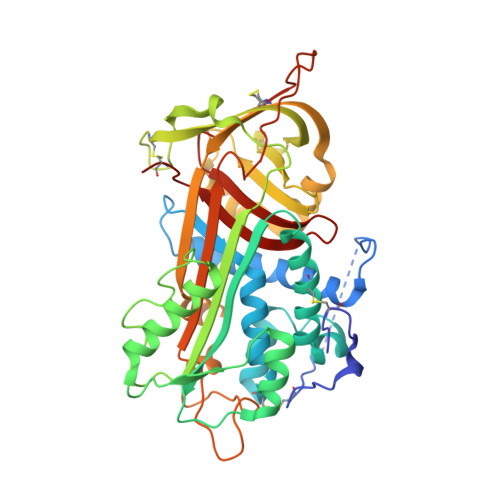
Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Li, W., Johnson, D.J., Esmon, C.T., Huntington, J.A.(2004) Nat Struct Mol Biol 11: 857-862
- PubMed: 15311269
- DOI: https://doi.org/10.1038/nsmb811
- Primary Citation of Related Structures:
1TB6 - PubMed Abstract:
The maintenance of normal blood flow depends completely on the inhibition of thrombin by antithrombin, a member of the serpin family. Antithrombin circulates at a high concentration, but only becomes capable of efficient thrombin inhibition on interaction with heparin or related glycosaminoglycans. The anticoagulant properties of therapeutic heparin are mediated by its interaction with antithrombin, although the structural basis for this interaction is unclear. Here we present the crystal structure at a resolution of 2.5 A of the ternary complex between antithrombin, thrombin and a heparin mimetic (SR123781). The structure reveals a template mechanism with antithrombin and thrombin bound to the same heparin chain. A notably close contact interface, comprised of extensive active site and exosite interactions, explains, in molecular detail, the basis of the antithrombotic properties of therapeutic heparin.
- University of Cambridge, Department of Haematology, Division of Structural Medicine, Thrombosis Research Unit, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 2XY, UK.
Organizational Affiliation: