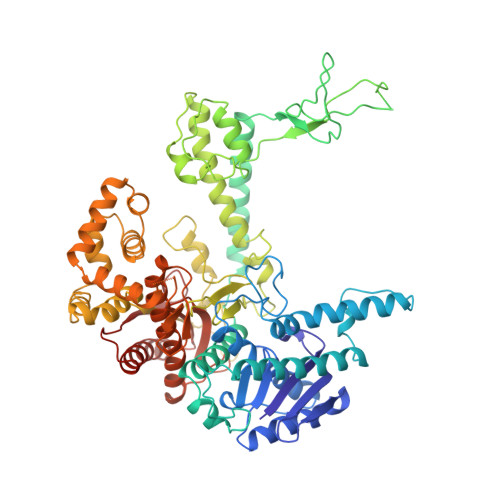
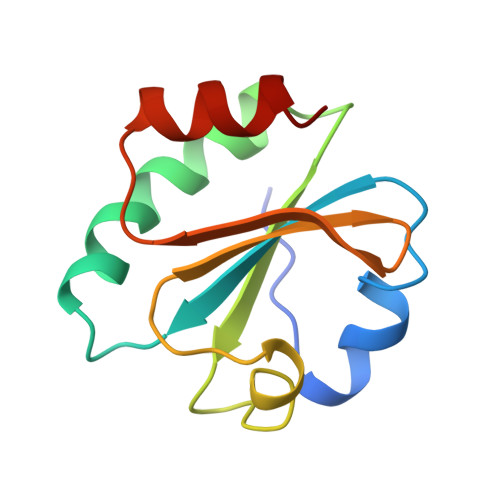
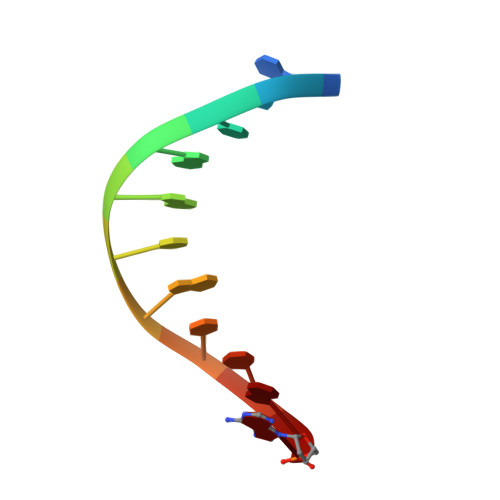
Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution.
Doublie, S., Tabor, S., Long, A.M., Richardson, C.C., Ellenberger, T.(1998) Nature 391: 251-258
- PubMed: 9440688
- DOI: https://doi.org/10.1038/34593
- Primary Citation of Related Structures:
1T7P - PubMed Abstract:
DNA polymerases change their specificity for nucleotide substrates with each catalytic cycle, while achieving error frequencies in the range of 10(-5) to 10(-6). Here we present a 2.2 A crystal structure of the replicative DNA polymerase from bacteriophage T7 complexed with a primer-template and a nucleoside triphosphate in the polymerase active site. The structure illustrates how nucleotides are selected in a template-directed manner, and provides a structural basis for a metal-assisted mechanism of phosphoryl transfer by a large group of related polymerases.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, USA.
Organizational Affiliation: