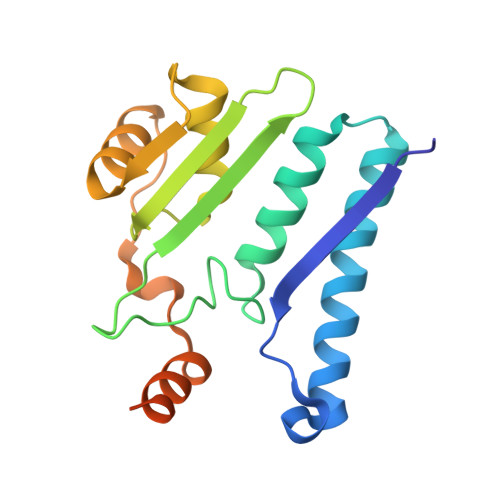
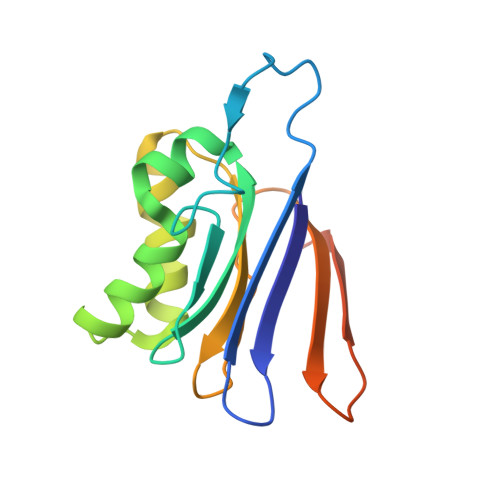
Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
Prahl, A., Pazgier, M., Hejazi, M., Lockau, W., Lubkowski, J.(2004) Acta Crystallogr D Biol Crystallogr 60: 1173-1176
- PubMed: 15159592
- DOI: https://doi.org/10.1107/S0907444904003403
- Primary Citation of Related Structures:
1T3M - PubMed Abstract:
The crystal structure of the Escherichia coli enzyme (EcAIII) with isoaspartyl dipeptidase and L-asparaginase activity has been solved and refined to a resolution of 1.65 angstroms, with crystallographic R-factor and Rfree values of 0.178 and 0.209, respectively. EcAIII belongs to the family of N-terminal hydrolases. The amino-acid sequence of EcAIII is homologous to those of putative asparaginases from plants. The structure of EcAIII is similar to the structures of glycosylasparaginases. The mature and catalytically active form of EcAIII is a heterotetramer consisting of two alpha-subunits and two beta-subunits. Both of the equivalent active sites present in the EcAIII tetramer is assisted by a metal-binding site. The metal cations, modelled here as Na+, have not previously been observed in glycosylasparaginases. This reported structure helps to explain the inability of EcAIII and other plant-type asparaginases to hydrolyze N4-(beta-N-acetylglucosaminyl)-L-asparagine, the substrate of glycosylasparaginases.
- Macromolecular Crystallography Laboratory, National Cancer Institute at Frederick, Frederick, MD 21702, USA.
Organizational Affiliation: