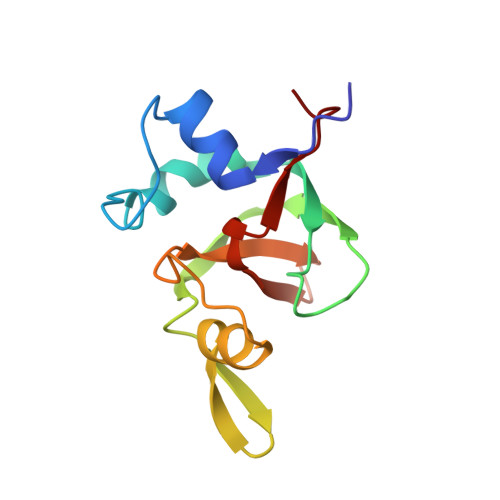
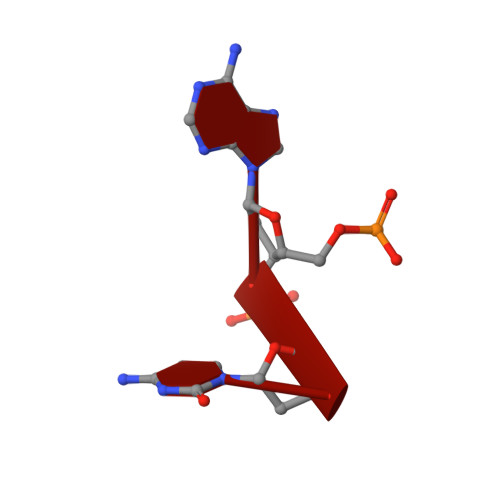
Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Lingel, A., Simon, B., Izaurralde, E., Sattler, M.(2004) Nat Struct Mol Biol 11: 576-577
- PubMed: 15156196
- DOI: https://doi.org/10.1038/nsmb777
- Primary Citation of Related Structures:
1T2R, 1T2S - PubMed Abstract:
We describe the solution structures of the Argonaute2 PAZ domain bound to RNA and DNA oligonucleotides. The structures reveal a unique mode of single-stranded nucleic acid binding in which the two 3'-terminal nucleotides are buried in a hydrophobic cleft. We propose that the PAZ domain contributes to the specific recognition of siRNAs by providing a binding pocket for their characteristic two-nucleotide 3' overhangs.
- European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.
Organizational Affiliation: