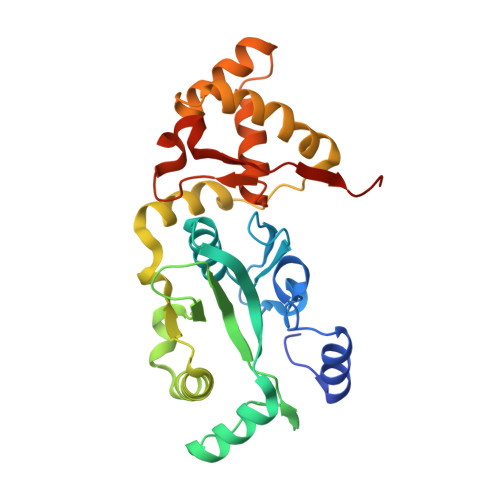
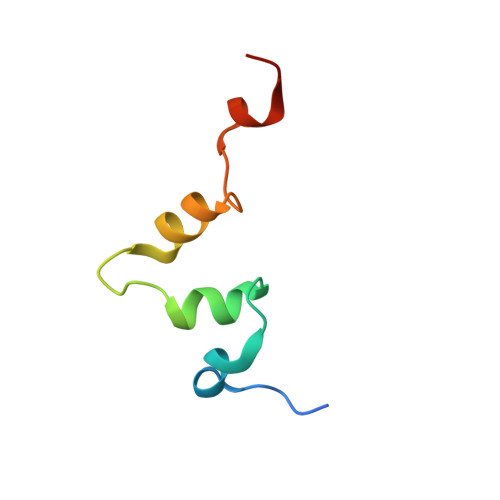
The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Ronning, D.R., Li, Y., Perez, Z.N., Ross, P.D., Hickman, A.B., Craig, N.L., Dyda, F.(2004) EMBO J 23: 2972-2981
- PubMed: 15257292
- DOI: https://doi.org/10.1038/sj.emboj.7600311
- Primary Citation of Related Structures:
1T0F - PubMed Abstract:
Tn7 transposition requires the assembly of a nucleoprotein complex containing four self-encoded proteins, transposon ends, and target DNA. Within this complex, TnsC, the molecular switch that regulates transposition, and TnsA, one part of the transposase, interact directly. Here, we demonstrate that residues 504-555 of TnsC are responsible for TnsA/TnsC interaction. The crystal structure of the TnsA/TnsC(504-555) complex, resolved to 1.85 A, illustrates the burial of a large hydrophobic patch on the surface of TnsA. One consequence of sequestering this patch is a marked increase in the thermal stability of TnsA as shown by differential scanning calorimetry. A model based on the complex structure suggested that TnsA and a slightly longer version of the cocrystallized TnsC fragment (residues 495-555) might cooperate to bind DNA, a prediction confirmed using gel mobility shift assays. Donor DNA binding by the TnsA/TnsC(495-555) complex is correlated with the activation of the TnsAB transposase, as measured by double-stranded DNA cleavage assays, demonstrating the importance of the TnsA/TnsC interaction in affecting Tn7 transposition.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD 20892, USA.
Organizational Affiliation: