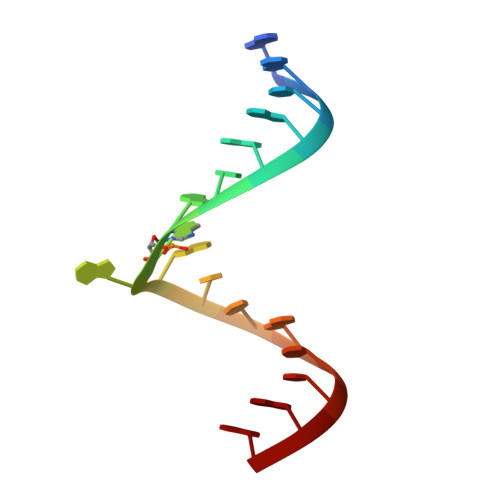
Monitoring molecular recognition of the ribosomal decoding site.
Shandrick, S., Zhao, Q., Han, Q., Ayida, B.K., Takahashi, M., Winters, G.C., Simonsen, K.B., Vourloumis, D., Hermann, T.(2004) Angew Chem Int Ed Engl 43: 3177-3182
- PubMed: 15199571
- DOI: https://doi.org/10.1002/anie.200454217
- Primary Citation of Related Structures:
1T0D, 1T0E - Department of Structural Chemistry, Anadys Pharmaceuticals, Inc 9050 Camino Santa Fe, San Diego, CA 92121, USA.
Organizational Affiliation: