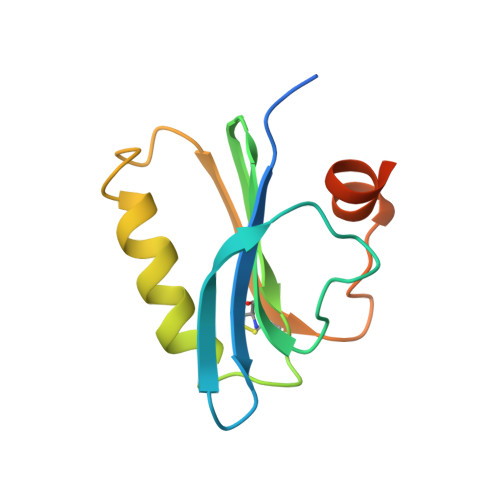
Structure of severin domain 2 in solution.
Schnuchel, A., Wiltscheck, R., Eichinger, L., Schleicher, M., Holak, T.A.(1995) J Mol Biology 247: 21-27
- PubMed: 7897658
- DOI: https://doi.org/10.1006/jmbi.1994.0118
- Primary Citation of Related Structures:
1SVQ, 1SVR - PubMed Abstract:
The three-dimensional structure of domain 2 of severin in aqueous solution was determined by nuclear magnetic resonance spectroscopy. Severin is a Ca(2+)-activated actin-binding protein that servers F-actin, nucleates actin assembly, and caps the fast-growing ends of actin filaments. The 114-residue domain consists of a central five-stranded beta-sheet, sandwiched between a parallel four-turn alpha-helix and, on the other face, a roughly perpendicular two-turn alpha-helix. There are two distinct binding sites for Ca2+ located near the N and C termini of the long helix. Conserved residues of the gelsolin-severin family contribute to the apolar core of domain 2 of severin, so that the overall fold of the protein is similar to those of segment 1 of gelsolin and profilins. Together with biochemical experiments, this structure helps to explain how severin interacts with actin.
- Max-Planck-Institute for Biochemistry, Martinsried, F.R.G.
Organizational Affiliation: