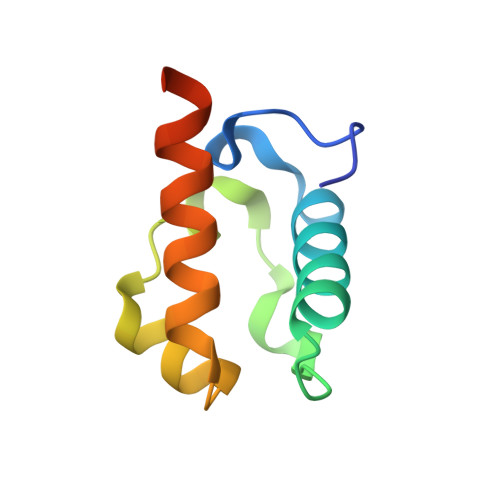
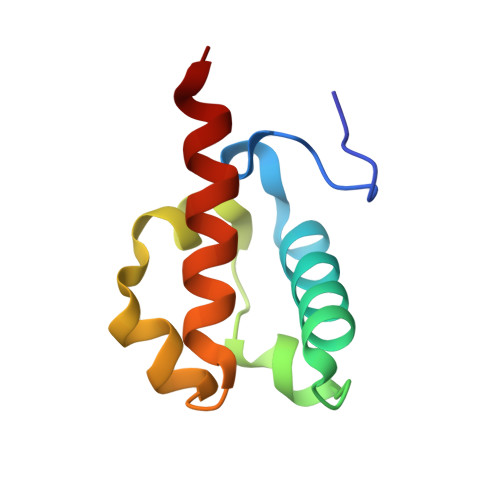
Derepression by depolymerization; structural insights into the regulation of yan by mae.
Qiao, F., Song, H., Kim, C.A., Sawaya, M.R., Hunter, J.B., Gingery, M., Rebay, I., Courey, A.J., Bowie, J.U.(2004) Cell 118: 163-173
- PubMed: 15260987
- DOI: https://doi.org/10.1016/j.cell.2004.07.010
- Primary Citation of Related Structures:
1SV0, 1SV4 - PubMed Abstract:
Yan, an ETS family transcriptional repressor, is regulated by receptor tyrosine kinase signaling via the Ras/MAPK pathway. Phosphorylation and downregulation of Yan is facilitated by a protein called Mae. Yan and Mae interact through their SAM domains. We find that repression by Yan requires the formation of a higher order structure mediated by Yan-SAM polymerization. Moreover, a crystal structure of the Yan-SAM/Mae-SAM complex shows that Mae-SAM specifically recognizes a surface on Yan-SAM that is also required for Yan-SAM polymerization. Mae-SAM binds to Yan-SAM with approximately 1000-fold higher affinity than Yan-SAM binds to itself and can effectively depolymerize Yan-SAM. Mutations on Mae that specifically disrupt its SAM domain-dependent interactions with Yan disable the derepression function of Mae in vivo. Depolymerization of Yan by Mae represents a novel mechanism of transcriptional control that sensitizes Yan for regulation by receptor tyrosine kinases.
- UCLA-DOE Institute of Genomics and Proteomics, Molecular Biology Institute, Department of Chemistry and Biochemistry, University of California, Los Angeles 90095, USA.
Organizational Affiliation: