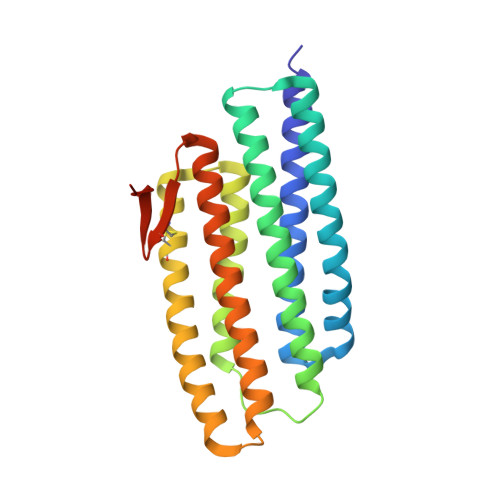
Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters.
Liu, J., Lou, Y., Yokota, H., Adams, P.D., Kim, R., Kim, S.H.(2005) J Biological Chem 280: 15960-15966
- PubMed: 15716271
- DOI: https://doi.org/10.1074/jbc.M414117200
- Primary Citation of Related Structures:
1SUM - PubMed Abstract:
PhoU proteins are known to play a role in the regulation of phosphate uptake. In Thermotoga maritima, two PhoU homologues have been identified bioinformatically. Here we report the crystal structure of one of the PhoU homologues at 2.0 A resolution. The structure of the PhoU protein homologue contains a highly symmetric new structural fold composed of two repeats of a three-helix bundle. The structure unexpectedly revealed a trinuclear and a tetranuclear iron cluster that were found to be bound on the surface. Each of the two multinuclear iron clusters is coordinated by a conserved E(D)XXXD motif pair. Our structure reveals a new class of metalloprotein containing multinuclear iron clusters. The possible functional implication based on the structure are discussed.
- Berkeley Structural Genomics Center, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.
Organizational Affiliation: