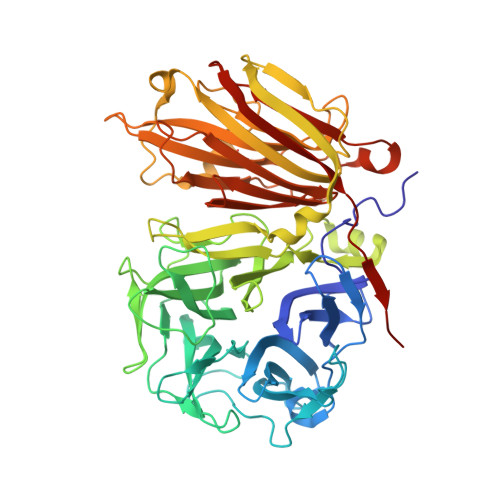
X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Verhaest, M., van den Ende, W., Roy, K.L., De Ranter, C.J., van Laere, A., Rabijns, A.(2005) Plant J 41: 400-411
- PubMed: 15659099
- DOI: https://doi.org/10.1111/j.1365-313X.2004.02304.x
- Primary Citation of Related Structures:
1ST8 - PubMed Abstract:
Fructan 1-exohydrolase, an enzyme involved in fructan degradation, belongs to the glycosyl hydrolase family 32. The structure of isoenzyme 1-FEH IIa from Cichorium intybus is described at a resolution of 2.35 A. The structure consists of an N-terminal fivefold beta-propeller domain connected to two C-terminal beta-sheets. The putative active site is located entirely in the beta-propeller domain and is formed by amino acids which are highly conserved within glycosyl hydrolase family 32. The fructan-binding site is thought to be in the cleft formed between the two domains. The 1-FEH IIa structure is compared with the structures of two homologous but functionally different enzymes: a levansucrase from Bacillus subtilis (glycosyl hydrolase family 68) and an invertase from Thermotoga maritima (glycosyl hydrolase family 32).
- Laboratorium voor Analytische Chemie en Medicinale Fysicochemie, Faculteit Farmaceutische Wetenschappen, K.U. Leuven, E. Van Evenstraat 4, B-3000 Leuven, Belgium.
Organizational Affiliation: