Insights into function of PSI domains from structure of the Met receptor PSI domain.
Kozlov, G., Perreault, A., Schrag, J.D., Park, M., Cygler, M., Gehring, K., Ekiel, I.(2004) Biochem Biophys Res Commun 321: 234-240
- PubMed: 15358240
- DOI: https://doi.org/10.1016/j.bbrc.2004.06.132
- Primary Citation of Related Structures:
1SSL - PubMed Abstract:
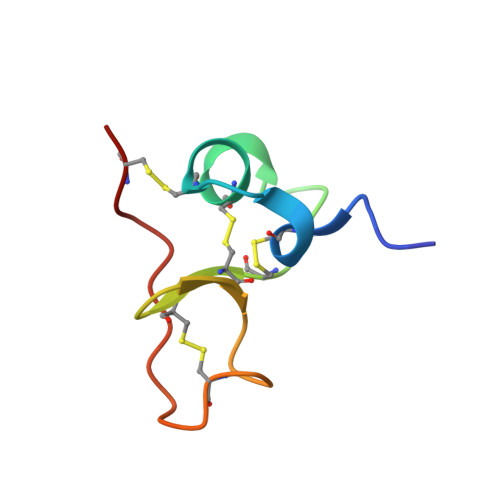
PSI domains are cysteine-rich modules found in extracellular fragments of hundreds of signaling proteins, including plexins, semaphorins, integrins, and attractins. Here, we report the solution structure of the PSI domain from the human Met receptor, a receptor tyrosine kinase critical for proliferation, motility, and differentiation. The structure represents a cysteine knot with short regions of secondary structure including a three-stranded antiparallel beta-sheet and two alpha-helices. All eight cysteines are involved in disulfide bonds with the pattern consistent with that for the PSI domain from Sema4D. Comparison with the Sema4D structure identifies a structurally conserved core comprising the N-terminal half of the PSI domain. Interestingly, this part links adjacent SEMA and immunoglobulin domains in the Sema4D structure, suggesting that the PSI domain serves as a wedge between propeller and immunoglobulin domains and is responsible for the correct positioning of the ligand-binding site of the receptor.
- Department of Biochemistry, McGill University, 3655 Promenade Sir William Osler, Montréal, Que., Canada H3G 1Y6.
Organizational Affiliation: