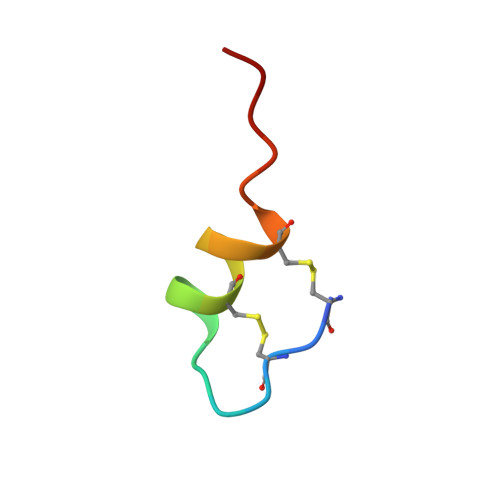
1H NMR studies of sarafotoxin SRTb, a nonselective endothelin receptor agonist, and IRL 1620, an ETB receptor-specific agonist.
Atkins, A.R., Martin, R.C., Smith, R.(1995) Biochemistry 34: 2026-2033
- PubMed: 7849060
- DOI: https://doi.org/10.1021/bi00006a024
- Primary Citation of Related Structures:
1SRB - PubMed Abstract:
1H NMR studies on the nonselective endothelin receptor agonist sarafotoxin SRTb have identified a helix between residues Asp 8 and His 16, and a beta-turn involving residues Cys 3 to Met 6; however, the biologically important C-terminal five residues were found to be conformationally variable. The average RMSD, measured for the final 43 refined structures to the average structure over residues 1-16, was 0.78 +/- 0.18 A for the backbone atoms and 1.39 +/- 0.22 A for all atoms. The torsion angles Cys 3 psi/Lys 4 theta, Thr 7 psi/Asp 8 theta and Gln 17 theta were identified as sites of conformational variability. Differences were found between the structures in the bicyclic loop region for SRTb and those published for ET1, another nonselective receptor agonist, which may explain the observed differences in potency of these peptides. The conformation of an ETB receptor-specific agonist, IRL 1620, which lacks the N-terminal seven residues and the two intrachain disulfides, was found by NMR and circular dichroism spectroscopy to be predominantly random coil, despite the fact that its affinity for the ETB receptor almost equals that of ET1. However, close analysis of the NMR results indicated the presence of turn-like structures, or a nascent helix, in the part of the sequence corresponding to the helical region in the parent peptides. These results suggest that the helical conformation may be required for ligand binding to the ETB receptor as well as to the ETA receptor.
- Biochemistry Department, University of Queensland, Australia.
Organizational Affiliation: