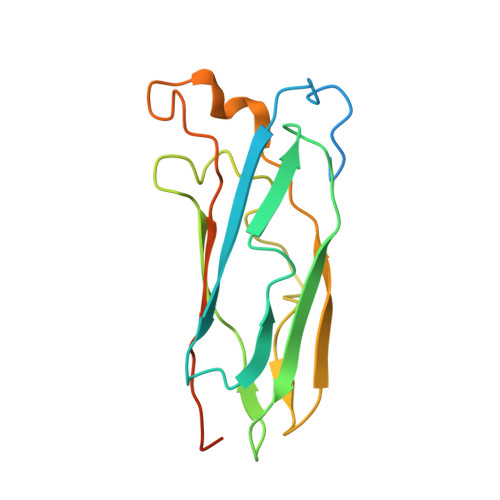
Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
Banci, L., Bertini, I., Cantini, F., Ciofi-Baffoni, S., Gonnelli, L., Mangani, S.(2004) J Biological Chem 279: 34833-34839
- PubMed: 15181013
- DOI: https://doi.org/10.1074/jbc.M403655200
- Primary Citation of Related Structures:
1SO9, 1SP0 - PubMed Abstract:
Cytochrome c oxidase assembly process involves many accessory proteins including Cox11, which is a copper-binding protein required for Cu incorporation into the Cu(B) site of cytochrome c oxidase. In a genome wide search, a number of Cox11 homologs are found in all of the eukaryotes with complete genomes and in several Gram-negative bacteria. All of them possess a highly homologous soluble domain and contain an N-terminal fragment that anchors the protein to the membrane. An anchor-free construct of 164 amino acids was obtained from Sinorhizobium meliloti, and the first structure of this class of proteins is reported here. The apoform has an immunoglobulin-like fold with a novel type of beta-strand organization. The copper binding motif composed of two highly conserved cysteines is located on one side of the beta-barrel structure. The apoprotein is monomeric in the presence of dithiothreitol, whereas it dimerizes in the absence of the reductant. When copper(I) binds, NMR and extended x-ray absorption fine structure (EXAFS) data indicate a dimeric protein state with two thiolates bridging two copper(I) ions. The present results advance the knowledge on the poorly understood molecular aspects of cytochrome c oxidase assembly.
- Magnetic Resonance Center CERM and Department of Chemistry, University of Florence, Via Luigi Sacconi 6, Sesto Fiorentino 50019, Florence, Italy.
Organizational Affiliation: