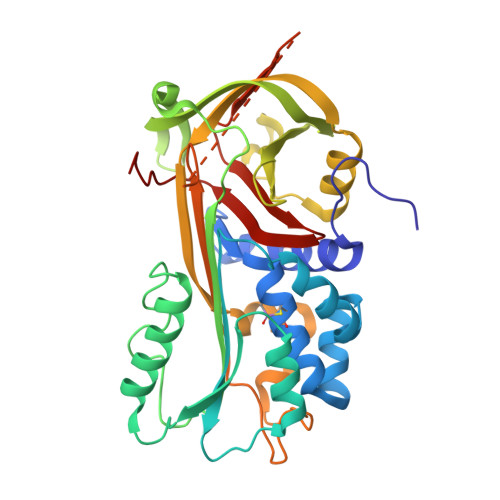
The high resolution crystal structure of a native thermostable serpin reveals the complex mechanism underpinning the stressed to relaxed transition.
Fulton, K.F., Buckle, A.M., Cabrita, L.D., Irving, J.A., Butcher, R.E., Smith, I., Reeve, S., Lesk, A.M., Bottomley, S.P., Rossjohn, J., Whisstock, J.C.(2005) J Biological Chem 280: 8435-8442
- PubMed: 15590653
- DOI: https://doi.org/10.1074/jbc.M410206200
- Primary Citation of Related Structures:
1SNG - PubMed Abstract:
Serpins fold into a native metastable state and utilize a complex conformational change to inhibit target proteases. An undesirable result of this conformational flexibility is that most inhibitory serpins are heat sensitive, forming inactive polymers at elevated temperatures. However, the prokaryote serpin, thermopin, from Thermobifida fusca is able to function in a heated environment. We have determined the 1.8 A x-ray crystal structure of thermopin in the native, inhibitory conformation. A structural comparison with the previously determined 1.5 A structure of cleaved thermopin provides detailed insight into the complex mechanism of conformational change in serpins. Flexibility in the shutter region and electrostatic interactions at the top of the A beta-sheet (the breach) involving the C-terminal tail, a unique structural feature of thermopin, are postulated to be important for controlling inhibitory activity and triggering conformational change, respectively, in the native state. Here we have discussed the structural basis of how this serpin reconciles the thermodynamic instability necessary for function with the stability required to withstand elevated temperatures.
- Protein Crystallography Unit, Monash Centre for Synchrotron Science, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Faculty of Medicine, Victorian Bioinformatics Consortium, P. O. Box 53, Australia.
Organizational Affiliation: