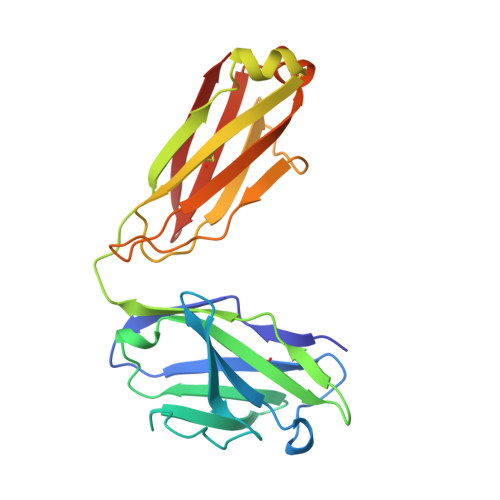
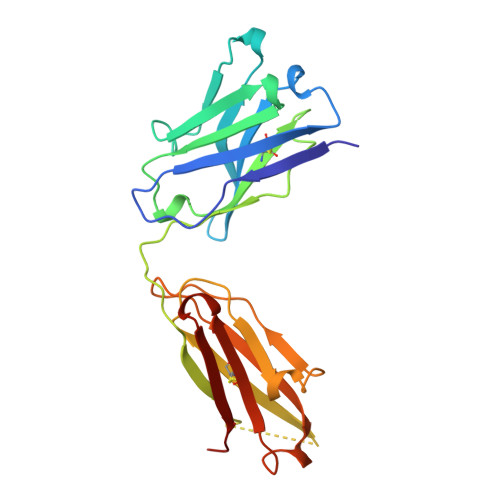
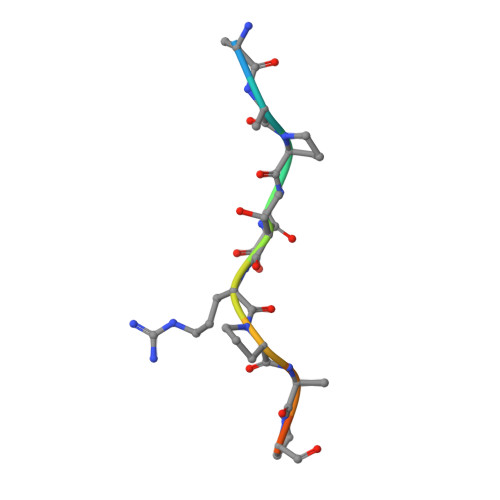
Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
Dokurno, P., Bates, P.A., Band, H.A., Stewart, L.M., Lally, J.M., Burchell, J.M., Taylor-Papadimitriou, J., Snary, D., Sternberg, M.J., Freemont, P.S.(1998) J Mol Biology 284: 713-728
- PubMed: 9826510
- DOI: https://doi.org/10.1006/jmbi.1998.2209
- Primary Citation of Related Structures:
1SM3 - PubMed Abstract:
The anti-breast tumour antibody SM3 has a high selectivity in reacting specifically with carcinoma-associated mucin. SM3 recognises the core repeating motif (Pro-Asp-Thr-Arg-Pro) of aberrantly glycosylated epithelial mucin MUC1, and has potential as a therapeutic and diagnostic tool. Here we report the crystal structure of the Fab fragment of SM3 in complex with a 13-residue MUC1 peptide antigen (Thr1P-Ser2P-Ala3P-Pro4P-Asp5P-Thr6P -Arg7P-Pro8P-Ala9P-Pro10P-Gly11P- Ser12P-Thr13P). The SM3-MUC1 peptide structure was solved by molecular replacement, and the current model is refined at 1.95 A resolution with an R-factor of 21.3% and R-free 28.3%. The MUC1 peptide is bound both by non-polar interactions and hydrogen bonds in an elongated groove in the antibody-combining site through interactions with Complimentarity Determining Regions (CDRs), three of the light chain (L1, L2, L3) and two of the heavy chain (H1 and H3). The conformation of the peptide is mainly extended with no discernable standard secondary structure. There is a single non-proline cis-peptide bond in H3 (Val95H-Gly96H-Gln97H-Phe98H-Ala101H-Ty r102H) between Gly96H and Gln97H, which appears to play a role in SM3-peptide antigen interactions, and represents the first such example within an antibody hypervariable loop. The SM3-MUC1 peptide structure has implications for rational therapeutic and diagnostic antibody engineering.
- Molecular Structure and Function Laboratory, Imperial Cancer Research Fund, London, UK.
Organizational Affiliation: