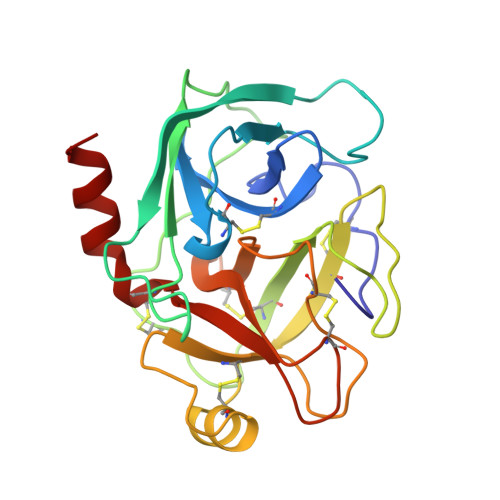
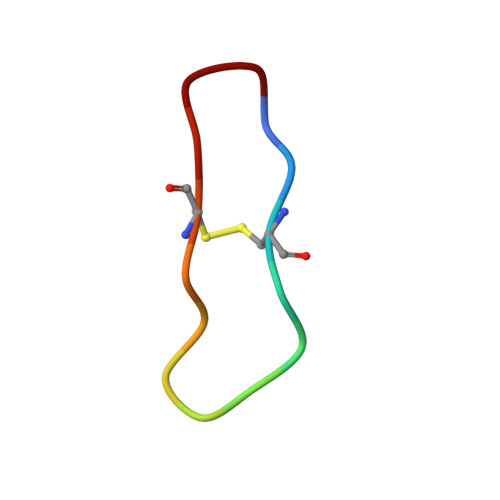
High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds.
Luckett, S., Garcia, R.S., Barker, J.J., Konarev, A.V., Shewry, P.R., Clarke, A.R., Brady, R.L.(1999) J Mol Biology 290: 525-533
- PubMed: 10390350
- DOI: https://doi.org/10.1006/jmbi.1999.2891
- Primary Citation of Related Structures:
1SFI - PubMed Abstract:
Proteinaceous serine proteinase inhibitors are widespread throughout the plant kingdom where they play an important role in protection against pests and pathogens. Here, we describe the isolation and characterisation of a novel 14 amino acid residue cyclic peptide from sunflower seeds, which is a potent inhibitor of trypsin (Ki=100 pM). The crystal structure of this peptide in complex with bovine beta-trypsin shows both sequence and conformational similarity with the trypsin-reactive loop of the Bowman-Birk family of serine proteinase inhibitors. This inhibitor, however, is unique in being monofunctional, cyclic and far shorter (14 amino acid residues) than inhibitors belonging to this family (typically 60-70 amino acid residues). The high potency of this peptide is likely to arise from the considerable structural rigidity achieved through its cyclic nature which is further stabilised by a single internal disulphide bond. This study helps delineate the minimal unit required for effective peptide inhibitors of serine proteinases, and will assist in the further design of inhibitors to this widespread class of enzymes.
- Department of Biochemistry, University of Bristol, Bristol, BS8 1TD, UK.
Organizational Affiliation: