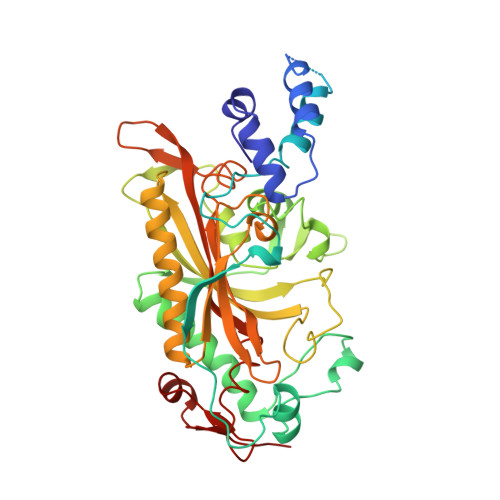
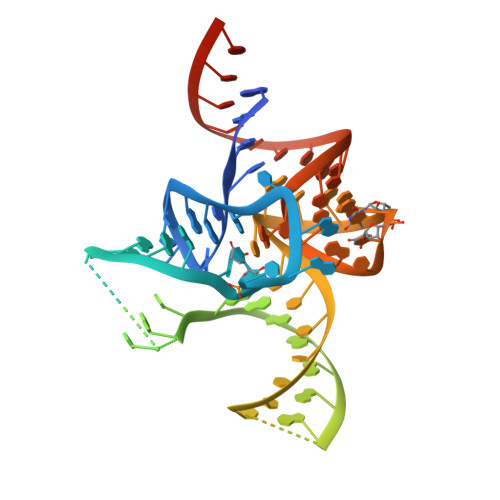
The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser).
Biou, V., Yaremchuk, A., Tukalo, M., Cusack, S.(1994) Science 263: 1404-1410
- PubMed: 8128220
- DOI: https://doi.org/10.1126/science.8128220
- Primary Citation of Related Structures:
1SER - PubMed Abstract:
The crystal structure of Thermus thermophilus seryl-transfer RNA synthetase, a class 2 aminoacyl-tRNA synthetase, complexed with a single tRNA(Ser) molecule was solved at 2.9 A resolution. The structure revealed how insertion of conserved base G20b from the D loop into the core of the tRNA determines the orientation of the long variable arm, which is a characteristic feature of most serine specific tRNAs. On tRNA binding, the antiparallel coiled-coil domain of one subunit of the synthetase makes contacts with the variable arm and T psi C loop of the tRNA and directs the acceptor stem of the tRNA into the active site of the other subunit. Specificity depends principally on recognition of the shape of tRNA(Ser) through backbone contacts and secondarily on sequence specific interactions.
- European Molecular Biology Laboratory, Grenoble Outstation, France.
Organizational Affiliation: