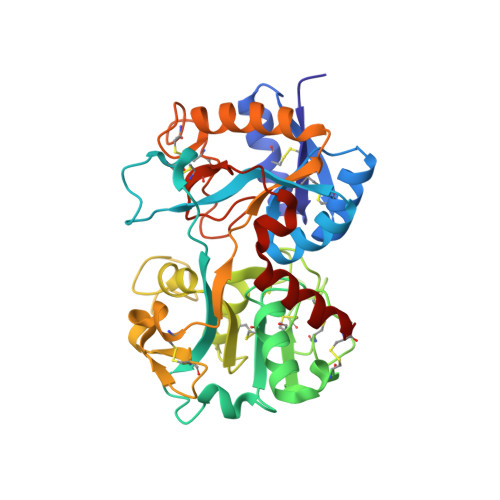
Structure of the zinc-saturated C-terminal lobe of bovine lactoferrin at 2.0 A resolution.
Jabeen, T., Sharma, S., Singh, N., Bhushan, A., Singh, T.P.(2005) Acta Crystallogr D Biol Crystallogr 61: 1107-1115
- PubMed: 16041076
- DOI: https://doi.org/10.1107/S0907444905016069
- Primary Citation of Related Structures:
1SDX - PubMed Abstract:
The crystal structure of the zinc-saturated C-terminal lobe of bovine lactoferrin has been determined at 2.0 A resolution using crystals stabilized at pH 3.8. This is the first metal-saturated structure of any functional lactoferrin at such a low pH. Purified samples of proteolytically generated zinc-saturated C-terminal lobe were crystallized from 0.1 M MES buffer pH 6.5 containing 25%(v/v) polyethyleneglycol monomethyl ether 550 and 0.1 M zinc sulfate heptahydrate. The crystals were transferred to 25 mM ammonium acetate buffer containing 25%(v/v) polyethyleneglycol monomethyl ether 550 and the pH was gradually changed from 6.5 to 3.8. The X-ray intensity data were collected with a 345 mm imaging-plate scanner mounted on an RU-300 rotating-anode X-ray generator using crystals soaked in the buffer at pH 3.8. The structure was determined with the molecular-replacement method using the coordinates of the monoferric C-terminal lobe of bovine lactoferrin as a search model and was refined to an R factor of 0.192 for all data to 2.0 A resolution. The final model comprises 2593 protein atoms (residues 342-676 and 681-685), 138 carbohydrate atoms (from 11 monosaccharide units in three glycan chains), three Zn2+ ions, one CO3(2-) ion, one SO(4)2- ions and 227 water molecules. The overall folding of the present structure is essentially similar to that of the monoferric C-terminal lobe of bovine lactoferrin, although it contains Zn2+ in place of Fe3+ in the metal-binding cleft as well as two additional Zn2+ ions on the surface of the C-terminal lobe. The Zn2+ ion in the cleft remains bound to the lobe with octahedral coordination. The bidentate carbonate ion is stabilized by a network of hydrogen bonds to Ala465, Gly466, Thr459 and Arg463. The other two zinc ions also form sixfold coordinations involving symmetry-related protein and water molecules. The number of monosaccharide residues from the three glycan chains of the C-terminal lobe was 11, which is the largest number observed to date. The structure shows that the C-terminal lobe of lactoferrin is capable of sequestering a Zn2+ ion at a pH of 3.8. This implies that the zinc ions can be sequestered over a wide pH range. The glycan chain attached to Asn545 may also have some influence on iron release from the C-terminal lobe.
- Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110 029, India.
Organizational Affiliation: