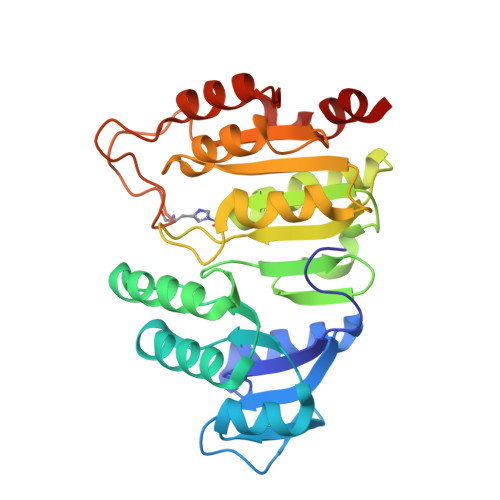
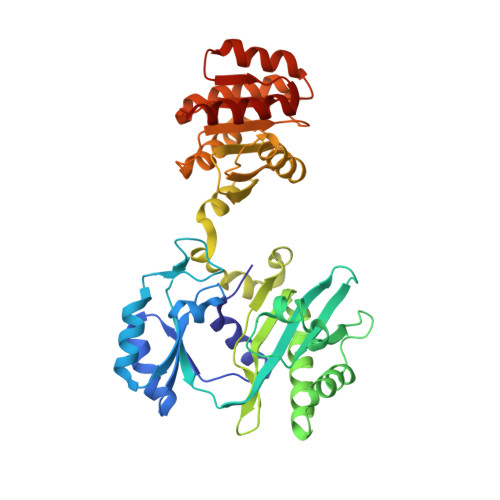
The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-A resolution.
Wolodko, W.T., Fraser, M.E., James, M.N., Bridger, W.A.(1994) J Biological Chem 269: 10883-10890
- PubMed: 8144675
- DOI: https://doi.org/10.2210/pdb1scu/pdb
- Primary Citation of Related Structures:
1SCU - PubMed Abstract:
The x-ray crystal structure of succinyl-CoA synthetase (SCS) from Escherichia coli has been determined by the method of multiple isomorphous replacement to a resolution of 2.5 A. Crystals of SCS are tetragonal with a space group of P4(3)22 and unit cell dimensions of a = b = 98.47 A and c = 400.6 A. One molecule of SCS (142 kDa) is contained in the asymmetric unit. The current model has been refined to a conventional R factor of 21.6% with root mean square deviations from ideal stereochemistry of 0.022 A for bond lengths and 3.25 degrees for bond angles. The quaternary organization of the E. coli enzyme is an alpha 2 beta 2 heterotetramer. In this tetramer, the alpha-subunits interact only with the beta-subunits, whereas the beta-subunits interact to form the dimer of alpha beta-dimers. The two active site pockets are located at regions of contact between alpha- and beta-subunits. One molecule of coenzyme A is bound to each alpha-subunit at a typical nucleotide-binding motif, and His-246 of each alpha-subunit is phosphorylated. This phosphohistidine, a catalytic intermediate, is stabilized by two helix dipoles (the "power" helices), one from each of the two subunit types. A short segment of the beta-subunit from one alpha beta-dimer is in close proximity to the CoA-binding site of the other alpha beta-dimer, providing a possible rationale for the overall tetrameric structure.
- Department of Biochemistry, University of Alberta, Edmonton, Canada.
Organizational Affiliation: