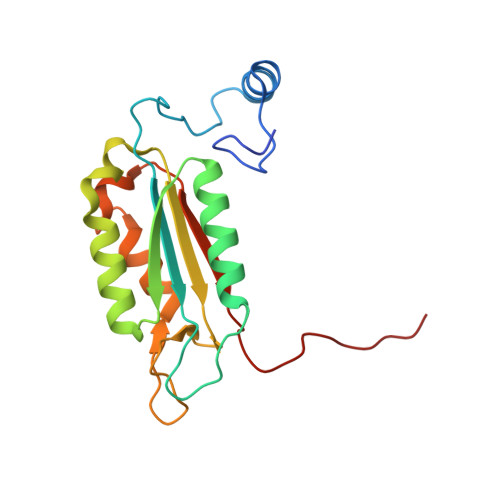
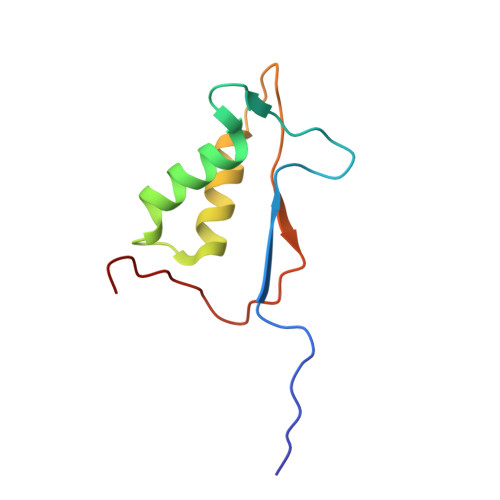
Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Romanowski, M.J., Scheer, J.M., O'Brien, T., McDowell, R.S.(2004) Structure 12: 1361-1371
- PubMed: 15296730
- DOI: https://doi.org/10.1016/j.str.2004.05.010
- Primary Citation of Related Structures:
1SC1, 1SC3, 1SC4 - PubMed Abstract:
Caspase-1, a mediator of the posttranslational processing of IL-1beta and IL-18, requires an aspartic acid in the P1 position of its substrates. The mechanisms of caspase-1 activation remain poorly understood despite numerous structures of the enzyme complexed with aspartate-based inhibitors. Here we report a crystal structure of ligand-free caspase-1 that displays dramatic rearrangements of loops defining the active site to generate a closed conformation that is incompatible with substrate binding. A structure of the enzyme complexed with malonate shows the protein in its open (active-site ligand-bound) conformation in which malonate reproduces the hydrogen bonding network observed in structures with covalent inhibitors. These results illustrate the essential function of the obligatory aspartate recognition element that opens the active site of caspase-1 to substrates and may be the determinant responsible for the conformational changes between ligand-free and -bound forms of the enzyme, and suggest a new approach for identifying novel aspartic acid mimetics.
- Department of Structural Biology, Sunesis Pharmaceuticals, Inc., 341 Oyster Point Boulevard, South San Francisco, California 94080, USA. romanom@sunesis.com
Organizational Affiliation: