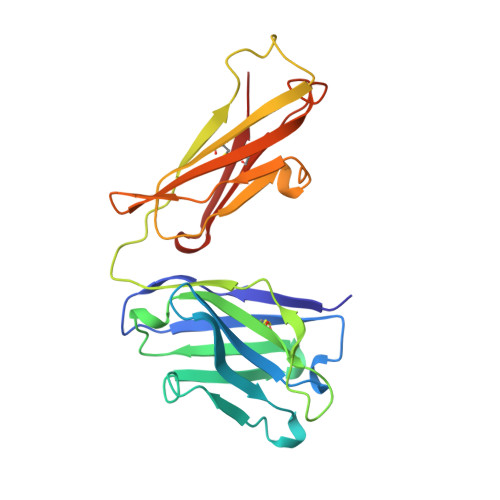
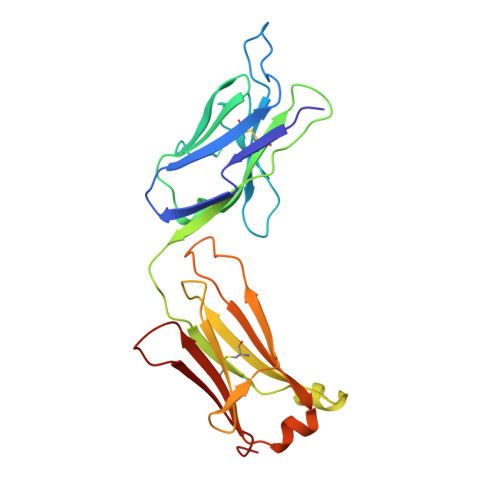
Structure of an Fab fragment against a C-terminal peptide of hCG at 2.0 A resolution.
Fotinou, C., Beauchamp, J., Emsley, P., deHaan, A., Schielen, W.J., Bos, E., Isaacs, N.W.(1998) J Biological Chem 273: 22515-22518
- PubMed: 9712877
- DOI: https://doi.org/10.1074/jbc.273.35.22515
- Primary Citation of Related Structures:
1SBS - PubMed Abstract:
3A2 is an antibody raised against human chorionic gonadotropin and recognizes a linear epitope on the C-terminal peptide of the human chorionic gonadotropin beta-subunit. Its three-dimensional structure has been determined to 2-A resolution using molecular replacement and refined to a conventional R-factor of 18.2%. The protein exhibits the typical immunoglobulin fold, and the model contains 944 ordered water molecules and one sulfate ion. A comparison of the complementarity-determining regions of the Fab3A2 with those from the Protein Data Bank following the canonical structure method reveals a canonical main chain conformation. This antibody belongs to the canonical structure class (combination of canonical conformations of the complementarity determining loops) that shows a preference for haptens and not for peptides. However, the shape of the surface of the antigen binding loops resembles that of an anti-peptide antibody.
- Department of Chemistry, University of Glasgow, Glasgow G12 8QQ, United Kingdom.
Organizational Affiliation: