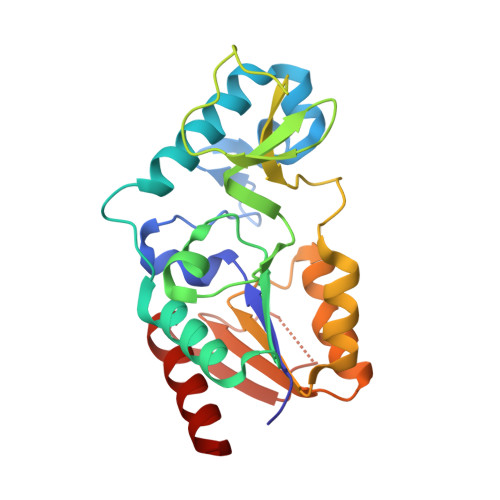
Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
Zhao, K., Chai, X., Marmorstein, R.(2004) J Mol Biology 337: 731-741
- PubMed: 15019790
- DOI: https://doi.org/10.1016/j.jmb.2004.01.060
- Primary Citation of Related Structures:
1S5P - PubMed Abstract:
Sirtuins are NAD+-dependent protein deacetylase enzymes that are broadly conserved from bacteria to human, and have been implicated to play important roles in gene regulation, metabolism and longevity. cobB is a bacterial sirtuin that deacetylates acetyl-CoA synthetase (Acs) at an active site lysine to stimulate its enzymatic activity. Here, we report the structure of cobB bound to an acetyl-lysine containing non-cognate histone H4 substrate. A comparison with the previously reported archaeal and eukaryotic sirtuin structures reveals the greatest variability in a small zinc-binding domain implicated to play a particularly important role in substrate-specific binding by the sirtuin proteins. Comparison of the cobB/histone H4 complex with other sirtuin proteins in complex with acetyl-lysine containing substrates, further suggests that contacts to the acetyl-lysine side-chain and beta-sheet interactions with residues directly C-terminal to the acetyl-lysine represent conserved features of sirtuin-substrate recognition. Isothermal titration calorimetry studies were used to compare the affinity of cobB for a variety of cognate and non-cognate acetyl-lysine-bearing peptides revealing an exothermic reaction with relatively little discrimination between substrates. In contrast, similar studies employing intact acetylated Acs protein as a substrate reveal a binding reaction that is endothermic, suggesting that cobB recognition of substrate involves a burial of hydrophobic surface and/or structural rearrangement involving substrate regions distal to the acetyl-lysine-binding site. Together, these studies suggest that substrate-specific binding by sirtuin proteins involves contributions from the zinc-binding domain of the enzyme and substrate regions distal to the acetyl-lysine-binding site.
- The Wistar Institute, University of Pennsylvania, Philadelphia, PA 19104, USA.
Organizational Affiliation: