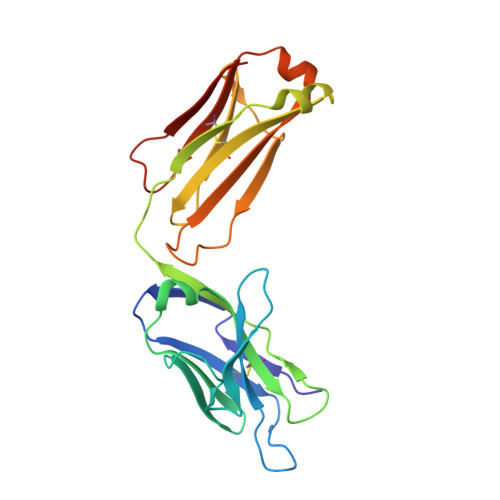
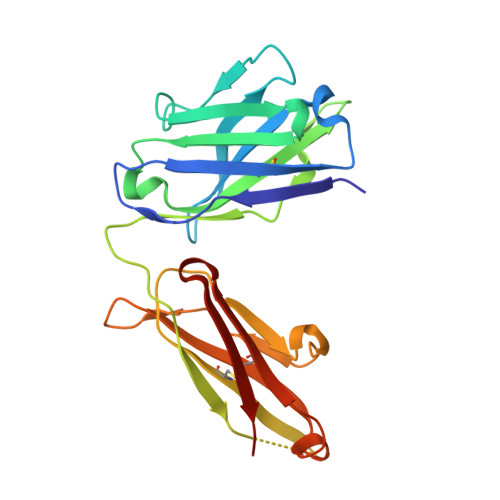
Structural convergence of antibody binding of carbohydrate determinants in lewis y tumor antigens
Ramsland, P.A., Farrugia, W., Bradford, T.M., Hogarth, P.M., Scott, A.M.(2004) J Mol Biology 340: 809-818
- PubMed: 15223322
- DOI: https://doi.org/10.1016/j.jmb.2004.05.037
- Primary Citation of Related Structures:
1S3K - PubMed Abstract:
Antibodies targeting human epithelial carcinomas bearing Lewis Y (Le(y)) carbohydrate antigens provide a striking illustration of convergent immune recognition. We report a 1.9A resolution crystal structure of the Fab of a humanized antibody (hu3S193) in complex with the Le(y) tetrasaccharide, Fuc(alpha 1-->2)Gal(beta 1-->4)[Fuc(alpha 1-->3)]GlcNAc. Comparisons of the hu3S193 and BR96 antibodies bound to Le(y) tumor antigens revealed extremely similar mechanisms for recognition of the carbohydrate epitopes. Solvent plays a critical role in hu3S193 antibody binding to the Le(y) carbohydrate epitope. Specificity for Le(y) is maintained because a conserved pocket accepts an N-acetyl group of the core Gal(beta 1-->4)GlcNAc disaccharide. Closely related blood-group determinants (Le(a) and Le(b)) cannot enter the specificity pocket, making the Le(y) antibodies promising candidates for immunotherapy of epithelial cancer.
- Structural Immunology Laboratory, Austin Research Institute, Heidelberg, Vic. 3084, Australia. p.ramsland@ari.unimelb.edu.au
Organizational Affiliation: