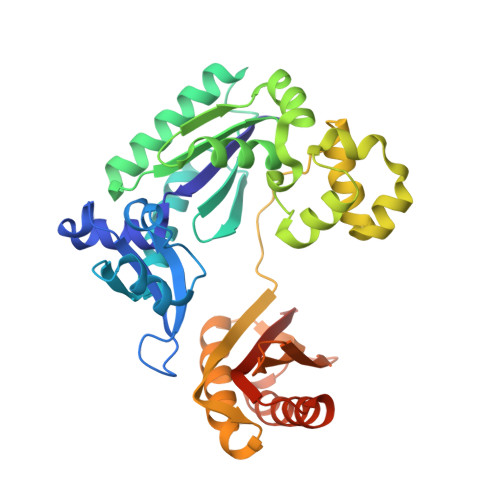
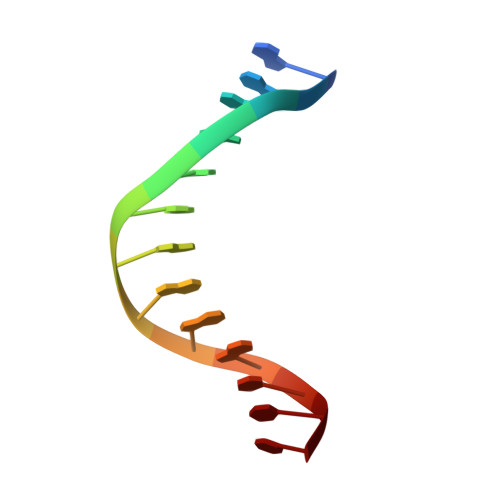
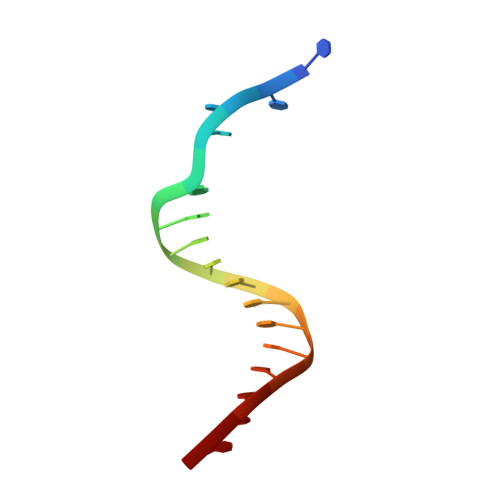
Snapshots of Replication through an Abasic Lesion; Structural Basis for Base Substitutions and Frameshifts.
Ling, H., Boudsocq, F., Woodgate, R., Yang, W.(2004) Mol Cell 13: 751-762
- PubMed: 15023344
- DOI: https://doi.org/10.1016/s1097-2765(04)00101-7
- Primary Citation of Related Structures:
1N56, 1S0N, 1S0O, 1S10 - PubMed Abstract:
Dpo4 from S. Solfataricus, a DinB-like Y family polymerase, efficiently replicates DNA past an abasic lesion. We have determined crystal structures of Dpo4 complexed with five different abasic site-containing DNA substrates and find that translesion synthesis is template directed with the abasic site looped out and the incoming nucleotide is opposite the base 5' to the lesion. The ensuing DNA synthesis generates a -1 frameshift when the abasic site remains extrahelical. Template realignment during primer extension is also observed, resulting in base substitutions or even +1 frameshifts. In the case of a +1 frameshift, the extra nucleotide is accommodated in the solvent-exposed minor groove. In addition, the structure of an unproductive Dpo4 ternary complex suggests that the flexible little finger domain facilitates DNA orientation and translocation during translesion synthesis.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: