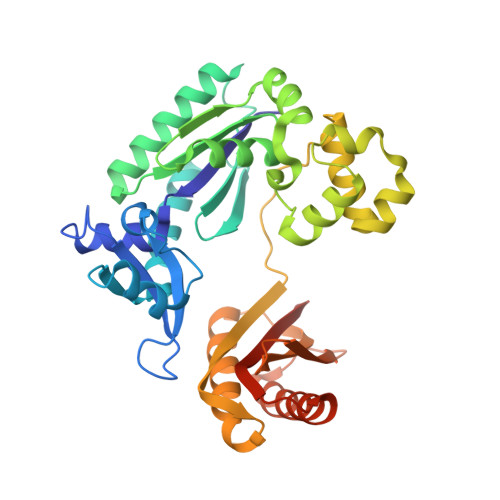
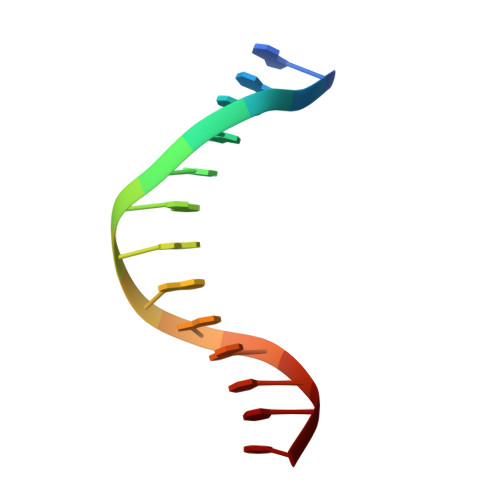
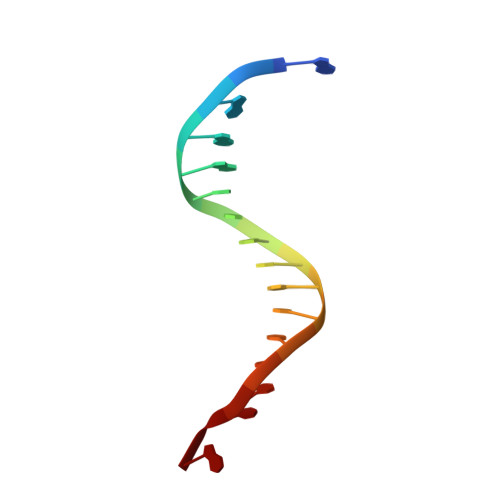
Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase.
Ling, H., Sayer, J.M., Plosky, B.S., Yagi, H., Boudsocq, F., Woodgate, R., Jerina, D.M., Yang, W.(2004) Proc Natl Acad Sci U S A 101: 2265-2269
- PubMed: 14982998
- DOI: https://doi.org/10.1073/pnas.0308332100
- Primary Citation of Related Structures:
1S0M - PubMed Abstract:
The first occupation-associated cancers to be recognized were the sooty warts (cancers of the scrotum) suffered by chimney sweeps in 18th century England. In the 19th century, high incidences of skin cancers were noted among fuel industry workers. By the early 20th century, malignant skin tumors were produced in laboratory animals by repeatedly painting them with coal tar. The culprit in coal tar that induces cancer was finally isolated in 1933 and determined to be benzo[a]pyrene (BP), a polycyclic aromatic hydrocarbon. A residue of fuel and tobacco combustion and frequently ingested by humans, BP is metabolized in mammals to benzo[a]pyrene diol epoxide (BPDE), which forms covalent DNA adducts and induces tumor growth. In the 70 yr since its isolation, BP has been the most studied carcinogen. Yet, there has been no crystal structure of a BPDE DNA adduct. We report here the crystal structure of a BPDE-adenine adduct base-paired with thymine at a template-primer junction and complexed with the lesion-bypass DNA polymerase Dpo4 and an incoming nucleotide. Two conformations of the BPDE, one intercalated between base pairs and another solvent-exposed in the major groove, are observed. The latter conformation, which can be stabilized by organic solvents that reduce the dielectric constant, seems more favorable for DNA replication by Dpo4. These structures also suggest a mechanism by which mutations are generated during replication of DNA containing BPDE adducts.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: