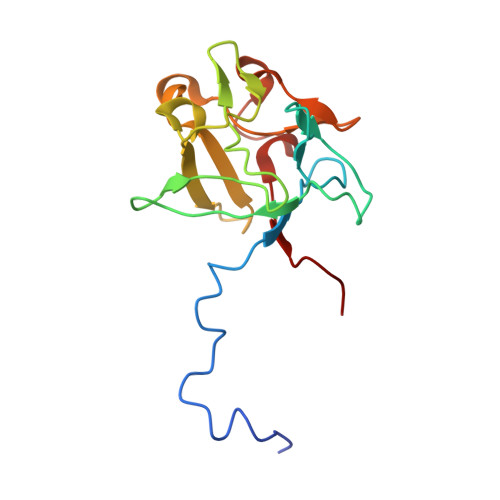
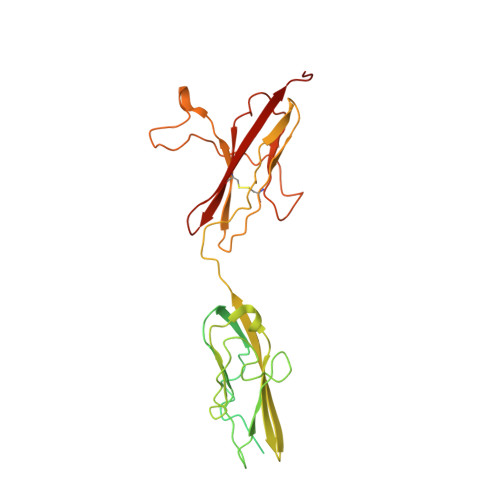
Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Olsen, S.K., Ibrahimi, O.A., Raucci, A., Zhang, F., Eliseenkova, A.V., Yayon, A., Basilico, C., Linhardt, R.J., Schlessinger, J., Mohammadi, M.(2004) Proc Natl Acad Sci U S A 101: 935-940
- PubMed: 14732692
- DOI: https://doi.org/10.1073/pnas.0307287101
- Primary Citation of Related Structures:
1RY7 - PubMed Abstract:
The prototypical fibroblast growth factor receptor (FGFR) extracellular domain consists of three Ig domains (D1-D3) of which the two membrane-proximal D2 and D3 domains and the interconnecting D2-D3 linker bear the determinants of ligand binding and specificity. In contrast, D1 and the D1-D2 linker are thought to play autoinhibitory roles in FGFR regulation. Here, we report the crystal structure of the three-Ig form of FGFR3c in complex with FGF1, an FGF that binds promiscuously to each of the seven principal FGFRs. In this structure, D1 and the D1-D2 linker are completely disordered, demonstrating that these regions are dispensable for FGF binding. Real-time binding experiments using surface plasmon resonance show that relative to two-Ig form, the three-Ig form of FGFR3c exhibits lower affinity for both FGF1 and heparin. Importantly, we demonstrate that this autoinhibition is mediated by intramolecular interactions of D1 and the D1-D2 linker with the minimal FGF and heparin-binding D2-D3 region. As in the FGF1-FGFR2c structure, but not the FGF1-FGFR1c structure, the alternatively spliced betaC'-betaE loop is ordered and interacts with FGF1 in the FGF1-FGFR3c structure. However, in contrast to the FGF1-FGFR2c structure in which the betaC'-betaE loop interacts with the beta-trefoil core region of FGF1, in the FGF1-FGFR3c structure, this loop interacts extensively with the N-terminal region of FGF1, underscoring the importance of the FGF1 N terminus in conferring receptor-binding affinity and promiscuity. Importantly, comparison of the three FGF1-FGFR structures shows that the flexibility of the betaC'-betaE loop is a major determinant of ligand-binding specificity and promiscuity.
- Departments of Pharmacology and Microbiology, New York University School of Medicine, New York, NY 10016, USA.
Organizational Affiliation: